- ▶
- Heaters/Source
- ▶
- Agilent Heaters and SensorsMass Spectrometry, Scientific Supplies & ManufacturingScientific Instrument Services 5973 Source Heater Tamper Resistant Allen Wrench 5973/5975 Quad Sensor 5985 Source Heater Assembly Agilent Interface Heater Assembly 5971 Interface Heater

- ▶
- Reference Material on InstrumentationArticle - A High Temperature Direct Probe for a Mass Spectrometer Design of a Direct Exposure Probe and Controller for use ona Hewlett-Packard 5989 Mass Spectrometer SIS AP1000 AutoProbe™ SIS AP2000 AutoProbe™ - Description of System HPP7: Direct Probe Electronics Console HPP7: Direct Probe for the Agilent (HP) 5973/5975 MSD HPP7: HP Direct Probe Application Notes HPP7: Installation Directions for the Direct Probe HPP7: Side Cover for the HP 5973 MSD HPP7: Support HPP7: Probe Inlet System for the Agilent (HP) 5973 and 5975 MSD with Automatic Indexed Stops HPP7: Theory of Operation of the Direct Probe and Probe Inlet System Direct Thermal Extraction Thermal Desorption Application Notes Environmental Thermal Desorption Application Notes Food Science Thermal Desorption Application Notes Forensic Thermal Desorption Application Notes GC Cryo-Trap Application Notes Headspace Application Notes Purge & Trap Thermal Desorption Application Notes Theory of Operation of the AutoDesorb® System AutoDesorb Notes for SIS Dealers Adsorbent Resin Application Notes Installation of the Short Path Thermal Desorption System on Agilent (HP) and Other GCs Installation of the Short Path Thermal Desorption System on a Varian 3400 GC AutoDesorb® System Development Team Thermal Desorption Applications and Reference Materials Installation of the Short Path Thermal Desorption System - TD5 Part I - Design & Operation of the Short Path ThermalDesorption System Installation Instructions for the Model 951 GC Cryo-Trap on the HP 5890 Series GC Installation Instructions for the Model 961 GC Cryo-Trap on the HP 5890 Series GC Operation of the Model 951/961 GC Cryo-Trap SIS GC Cryo Traps - Theory of Operation NIST/EPA/NIH Mass Spectral Enhancements - 1998 version (NIST98) SIMION 3D Ion Optics Class Mass Spectrometer Source Cleaning Methods MS Tip: Mass Spectrometer Source Cleaning Procedures Mass Spec Source Cleaning Procedures Micro-Mesh® Abrasive Sheets Research Papers Using New Era Syringe Pump Systems EI Positive Ion Spectra for Perfluorokerosene (PFK) Cap Liner Information How do I convert between fluid oz and milliliters? Which bottle material should I choose? Which bottle mouth should I choose? The Bottle Selection Guide CGA Connections for Gas Tanks Chemical Reaction Interface Mass Spectrometry (CRIMS)

- LiteratureApplication Notes Adsorbent Resins Guide Mass Spec Tips SDS Sheets FAQ MS Calibration Compound Spectra Manuals MS Links/Labs/ Organizations MS Online Tools Flyers on Products/Services Scientific Supplies Catalog About Us NextAdvance Bullet Blender® Homogenizer Protocols Micro-Mesh® Literature Instrumentation Literature Agilent GC/MS Literature SIS News / E-Mail Newsletter NIST MS Database - Update Notifications

- ▶
- Chemical Reaction Interface Mass Spectrometry (CRIMS)51. Development and Characterization of a New Chemical Interface for the Detection of Nonradioisotopically Labeled Analytes Using Mass Spectrometry (CRIMS) (EAS 96) 67. Using Chemical Reaction Interface (CRIMS) to Monitor Bacteria Transport in Situ (PittCon 98) 68. Using a Plug In UV-Vis Spectrometer to Monitor the Plasma Conditions in a GC CRIMS (EAS 98) 76. Determination of the Sensitivity of a CRIMS System (ASMS 98)

- Application NotesNote 103: EPA Method 325B, Novel Thermal Desorption Instrument Modification to Improve Sensitivity Note 102: Identification of Contaminants in Powdered Beverages by Direct Extraction Thermal Desorption GC/MS Note 101: Identification of Contaminants in Powdered Foods by Direct Extraction Thermal Desorption GC/MS Note 100: Volatile and Semi-Volatile Profile Comparison of Whole Versus Cracked Versus Dry Homogenized Barley Grains by Direct Thermal Extraction Note 99: Volatile and Semi-Volatile Profile Comparison of Whole vs. Dry Homogenized Wheat, Rye and Barley Grains by Direct Thermal Extraction GC/MS Note 98: Flavor and Aroma Profiles of Truffle Oils by Thermal Desorption GC/MS Note 97: Flavor Profiles of Imported and Domestic Beers by Purge & Trap Thermal Desorption GC/MS Note 96: Reducing Warping in Mass Spectrometer Filaments, with SISAlloy® Yttria/Rhenium Filaments Note 95: Detection of Explosives on Clothing Material by Direct and AirSampling Thermal Desorption GC/MS Note 94: Detection of Nepetalactone in the Nepeta Cataria Plant by Thermal Desorption GC/MS Note 93: Detection of Benzene in Carbonated Beverages with Purge & Trap Thermal Desorption GC/MS Note 92: Yttria Coated Mass Spectrometer Filaments Note 91: AutoProbe DEP Probe Tip Temperatures Note 90: An Automated MS Direct Probe for use in an Open Access Environment Note 89: Quantitation of Organics via a Mass Spectrometer Automated Direct Probe Note 88: Analysis of Silicone Contaminants on Electronic Components by Thermal Desorption GC-MS Note 87: Design and Development of an Automated Direct Probe for a Mass Spectrometer Note 86: Simulation of a Unique Cylindrical Quadrupole Mass Analyzer Using SIMION 7.0. Note 85: Replacing an Electron Multiplier in the Agilent (HP) 5973 MSD Note 84: Vacuum Pump Exhaust Filters - Charcoal Exhaust Traps Note 83: Vacuum Pump Exhaust Filters - Oil Mist Eliminators Note 82: Vacuum Pump Exhaust Filters Note 81: Rapid Bacterial Chemotaxonomy By DirectProbe/MSD Note 80: Design, Development and Testing of a Microprocessor ControlledAutomated Short Path Thermal Desorption Apparatus Note 79: Volatile Organic Compounds From Electron Beam Cured and Partially Electron Beam Cured Packaging Using Automated Short Path Thermal Desorption Note 78: A New Solution to Eliminate MS Down-Time With No-Tool-Changing of Analytical GC Columns Note 77: The Determination of Volatile Organic Compounds in VacuumSystem Components Note 76: Determination of the Sensitivity of a CRIMS System Note 75: An Apparatus for Sampling Volatile Organics From LivePlant Material Using Short Path Thermal Desorption Note 74: Examination of Source Design in Electrospray-TOF Using SIMION 3D Note 73: The Analysis of Perfumes and their Effect on Indoor Air Pollution Note 72: 1998 Version of the NIST/EPA/NIH Mass Spectral Library, NIST98 Note 71: Flavor Profile Determination of Rice Samples Using Shor tPath Thermal Desorption GC Methods Note 70: Application of SIMION 6.0 To a Study of the Finkelstein Ion Source: Part II Note 69: Application of SIMION 6.0 To a Study of the Finkelstein Ion Source: Part 1 Note 68: Use of a PC Plug-In UV-Vis Spectrometer To Monitor the Plasma Conditions In GC-CRIMS Note 67: Using Chemical Reaction Interface Mass Spectrometry (CRIMS) To Monitor Bacterial Transport In In Situ Bioremediation Note 66: Probe Tip Design For the Optimization of Direct Insertion Probe Performance Note 65: Determination of Ethylene by Adsorbent Trapping and Thermal Desorption - Gas Chromatography Note 64: Comparison of Various GC/MS Techniques For the Analysis of Black Pepper (Piper Nigrum) Note 63: Determination of Volatile and Semi-Volatile Organics in Printer Toners Using Thermal Desorption GC Techniques Note 62: Analysis of Polymer Samples Using a Direct Insertion Probe and EI Ionization Note 61: Analysis of Sugars Via a New DEP Probe Tip For Use With theDirect Probe On the HP5973 MSD Note 60: Programmable Temperature Ramping of Samples Analyzed ViaDirect Thermal Extraction GC/MS Note 59: Computer Modeling of a TOF Reflectron With Gridless Reflector Using SIMION 3D Note 58: Direct Probe Analysis and Identification of Multicomponent Pharmaceutical Samples via Electron Impact MS Note 57: Aroma Profiles of Lavandula species Note 56: Mass Spec Maintenance & Cleaning Utilizing Micro-Mesh® Abrasive Sheets Note 55: Seasonal Variation in Flower Volatiles Note 54: Identification of Volatile Organic Compounds in Office Products Note 53: SIMION 3D v6.0 Ion Optics Simulation Software Note 52: Computer Modeling of Ion Optics in Time-of-Flight mass Spectrometry Using SIMION 3D Note 51: Development and Characterization of a New Chemical Reaction Interface for the Detection of Nonradioisotopically Labeled Analytes Using Mass Spectrometry (CRIMS) Note 50: The Analysis of Multiple Component Drug Samples Using a Direct Probe Interfaced to the HP 5973 MSD Note 49: Analysis of Cocaine Utilizing a New Direct Insertion Probe on a Hewlett Packard 5973 MSD Note 48: Demonstration of Sensitivity Levels For the Detection of Caffeine Using a New Direct Probe and Inlet for the HP 5973 MSD Note 47: The Application Of SIMION 6.0 To Problems In Time-of-Flight Mass Spectrometry Note 46: Delayed Extraction and Laser Desorption: Time-lag Focusing and Beyond Note 45: Application of SIMION 6.0 to Filament Design for Mass Spectrometer Ionization Sources Note 44: The Design Of a New Direct Probe Inlet For a Mass Spectrometer Note 43: Volatile Organic Composition In Blueberries Note 42: The Influence of Pump Oil Purity on Roughing Pumps Note 41: Hydrocarbon Production in Pine by Direct Thermal Extraction Note 40: Comparison of Septa by Direct Thermal Extraction Note 39: Comparison of Sensitivity Of Headspace GC, Purge and Trap Thermal Desorption and Direct Thermal Extraction Techniques For Volatile Organics Note 38: A New Micro Cryo-Trap For Trapping Of Volatiles At the Front Of a GC Capillary Column Note 37: Volatile Organic Emissions from Automobile Tires Note 36: Identification Of Volatile Organic Compounds In a New Automobile Note 35: Volatile Organics Composition of Cranberries Note 34: Selection Of Thermal Desorption and Cryo-Trap Parameters In the Analysis Of Teas Note 33: Changes in Volatile Organic Composition in Milk Over Time Note 32: Selection and Use of Adsorbent Resins for Purge and Trap Thermal Desorption Applications Note 31: Volatile Organic Composition in Several Cultivars of Peaches Note 30: Comparison Of Cooking Oils By Direct Thermal Extraction and Purge and Trap GC/MS Note 29: Analysis Of Volatile Organics In Oil Base Paints By Automated Headspace Sampling and GC Cryo-Focusing Note 28: Analysis Of Volatile Organics In Latex Paints By Automated Headspace Sampling and GC Cryo-Focusing Note 27: Analysis of Volatile Organics In Soils By Automated Headspace GC Note 26: Volatile Organics Present in Recycled Air Aboard a Commercial Airliner Note 25: Flavor and Aroma in Natural Bee Honey Note 24: Selection of GC Guard Columns For Use With the GC Cryo-Trap Note 23: Frangrance Qualities in Colognes Note 22: Comparison Of Volatile Compounds In Latex Paints Note 21: Detection and Identification Of Volatile and Semi-Volatile Organics In Synthetic Polymers Used In Food and Pharmaceutical Packaging Note 20: Using Direct Thermal Desorption to Assess the Potential Pool of Styrene and 4-Phenylcyclohexene In Latex-Backed Carpets Note 19: A New Programmable Cryo-Cooling/Heating Trap for the Cryo-Focusing of Volatiles and Semi-Volatiles at the Head of GC Capillary Columns Note 18: Determination of Volatile Organic Compounds In Mushrooms Note 17: Identification of Volatile Organics in Wines Over Time Note 16: Analysis of Indoor Air and Sources of Indoor Air Contamination by Thermal Desorption Note 14: Identification of Volatiles and Semi-Volatiles In Carbonated Colas Note 13: Identification and Quantification of Semi-Volatiles In Soil Using Direct Thermal Desorption Note 12: Identification of the Volatile and Semi-Volatile Organics In Chewing Gums By Direct Thermal Desorption Note 11: Flavor/Fragrance Profiles of Instant and Ground Coffees By Short Path Thermal Desorption Note 10: Quantification of Naphthalene In a Contaminated Pharmaceutical Product By Short Path Thermal Desorption Note 9: Methodologies For the Quantification Of Purge and Trap Thermal Desorption and Direct Thermal Desorption Analyses Note 8: Detection of Volatile Organic Compounds In Liquids Utilizing the Short Path Thermal Desorption System Note 7: Chemical Residue Analysis of Pharmaceuticals Using The Short Path Thermal Desorption System Note 6: Direct Thermal Analysis of Plastic Food Wraps Using the Short Path Thermal Desorption System Note 5: Direct Thermal Analysis Using the Short Path Thermal Desorption System Note 4: Direct Analysis of Spices and Coffee Note 3: Indoor Air Pollution Note 2: Detection of Arson Accelerants Using Dynamic Headspace with Tenax® Cartridges Thermal Desorption and Cryofocusing Note 1: Determination of Off-Odors and Other Volatile Organics In Food Packaging Films By Direct Thermal Analysis-GC-MS Tech No. "A" Note 14: Elimination of "Memory" Peaks in Thermal Desorption Improving Sensitivity in the H.P. 5971 MSD and Other Mass Spectrometers - Part I of II Improving Sensitivity in the H.P. 5971 MSD and Other Mass Spectrometers- Part II of II Adsorbent Resins Guide Development and Field Tests of an Automated Pyrolysis Insert for Gas Chromatography. Hydrocarbon Production in Pine by Direct Thermal Extraction A New Micro Cryo-Trap for the Trapping of Volatiles at the Front of a GC Capillary (019P) - Comparison of Septa by Direct Thermal Extraction Volatile Organic Composition in Blueberry Identification of Volatile Organic Compounds in Office Products Detection and Indentification of Volatiles in Oil Base Paintsby Headspace GC with On Column Cryo-Trapping Evaluation of Septa Using a Direct Thermal Extraction Technique INFLUENCE OF STORAGE ON BLUEBERRY VOLATILES Selection of Thermal Desorption and Cryo-Trap Parameters in the Analysis of Teas Redesign and Performance of a Diffusion Based Solvent Removal Interface for LC/MS The Design of a New Direct Probe Inlet for a Mass Spectrometer Analytes Using Mass Spectrometry (CRIMS) Application of SIMION 6.0 to Filament Design for Mass Spectrometer Ionization Sources A Student Guide for SIMION Modeling Software Application of SIMION 6.0 to Problems in Time-of-flight Mass Spectrometry Comparison of Sensitivity of Headspace GC, Purge and TrapThermal Desorption and Direct Thermal Extraction Techniques forVolatile Organics The Influence of Pump Oil Purity on Roughing Pumps Analysis of Motor Oils Using Thermal Desorption-Gas Chromatography-Mass Spectrometry IDENTIFICATION OF VOLATILE ORGANIC COMPOUNDS IN PAPER PRODUCTS Computer Modeling of Ion Optics in Time-of-Flight mass Spectrometry using SIMION 3D Seasonal Variation in Flower Volatiles Development of and Automated Microprocessor Controlled Gas chromatograph Fraction Collector / Olfactometer Delayed Extraction and Laser Desorption: Time-lag Focusing and Beyond A New Micro Cryo-Trap for the Trapping of Volatiles at the Front of a GC Column Design of a Microprocessor Controlled Short Path Thermal Desorption Autosampler Computer Modeling of Ion Optics in Time-of-Flight Mass Spectrometry Using SIMION 3D Thermal Desorption Instrumentation for Characterization of Odors and Flavors

- ▶
- 67. Using Chemical Reaction Interface (CRIMS) to Monitor Bacteria Transport in Situ (PittCon 98) (This Page)
By Steven Colby, Eric Butrym, Scientific Instrument Services, Ringoes, NJ
By Tullis Onstott, Department of Geosciences, Princeton University, Princeton, NJ 08544.
Presented at PittCon 98, Orlando, FL., March 1998
Abstract
The introduction of degradative bacteria into aquifers is becoming an increasingly attractive bioremediation strategy. This is in part due to a significant cost advantage over traditional technologies. Obtaining a uniform bacterial distribution throughout the contaminated zone is pivotal to achieving rapid degradation rates. Tracking the migration of the bacteria injected underground is, therefore, of great interest to those involved in remediation of subsurface environments. Distinguishing the injected bacteria from the indigenous microbial population is often a difficult, time-consuming and hence an expensive process. Tagging the degradative microorganisms with a chemical or isotopic marker is a less costly alternative. This tactic can lead to erroneous results, however, if the marker becomes dissociated from the bacteria through death or predation.
Both of these problems are solved through the use of stable isotope labeling and detection of bacterial-specific lipids. This can be accomplished with the use of Gas Chromatography and Chemical Reaction Interface Mass Spectrometry (GC-CRIMS). The CRIMS technique identifies isotopically labeled species by isotope ratio determination. Chromatographic effluent is mixed with a reactant gas and passed into a microwave plasma. Specific combustion products are produced which permit the measurement of isotope intensities with the interference of isotopologs. This technique provides rapid quantification of the degradative bacterial concentration in subsurface samples at concentrations levels below the detection limit of conventional methods.
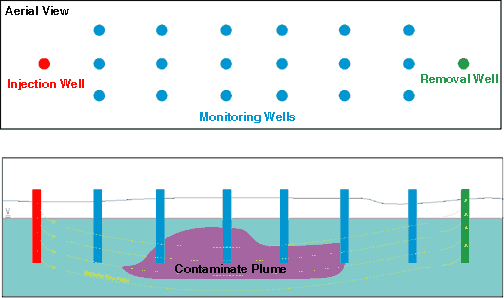
Figure 1 - Schematic representation of a typical bacterial injection for bioremediation of groundwater pollutants. Degradataive bacteria are pumped into the ground water at several depths below the water table via an injection well (red), ofaten mixed with nutrients and air. Tahe bacteria are carried througn conductive areas of the acquifer into the contaminated area (magenta) by the pushing action of the injection well, as well as the pulling action of the removal well(gareen). Monitaoring wells (blue) are used to remove samples for assessing the degradation and movement of the contaminant, as well as the effectiveness of bacterial transport through the contaminated area.
Introduction To Bioremediation and Bacterial Transport
Bioremediation is the use of biological processes to transform and degrade pollutants in the environment. Often times, this is accomplished by developing specific strains of bacteria which are highly effective at metabolizing the target pollutant, and are often engineered to possess other desirable traits as well. One of these desirable traits is a reduced tendency to adhere to sediment surfaces, and hence, a greater ability to be transported to the areas of the subsurface which are contaminated. In order to test potential organisms and monitor the movement of injected organisms during bioremediation activities, it is necessary to "tag" the degradative organisms is some way. This had traditionally been done by engineering the organisms to have a unique antibiotic sensitivity pattern. This method is falling out of favor, however, due to the rise in multiple antibiotic resistant pathogenic bacteria. In the laboratory, another dominant method has been to label the degradative organisms with a radioisotope (14C, 35S, 3H) and measure the radioactivity in the effluent from sediment cores, which allows very small numbers of organisms to be detected. For obvious reasons, this is not an acceptable method for field studies. The use of stable isotopes and GC-CRIMS, however, parallels the use of radioisotopes, and is expected to yield comparable results. When only specific components of degradative bacterial cells (ie., proteins, membrane fatty acids, DNA, RNA) are extracted and analyzed, it will also be possible to verify that any stable isotope observed represents the target organism, rather than a transfer of isotope from the injected bacteria to an indigenous bacteria or protozoa due to cell death or predation.
Introduction To CRIMS
Chemical Reaction Interface/Mass Spectrometry is a selective, sensitive, and versatile technique by which specific isotopes or elements can be monitored. CRIMS parallels the use of radioisotopes in that each technique monitors for elemental tags that are independent of the tags chemical structure or environment. In the CRIMS technique, chromatography effluent is introduced into a high-temperature microwave plasma. The analyte molecules are decomposed and then reacted with a specific reactant gas. This results in a small number of well characterized polyatomic products. These compounds are then detected by mass spectrometry. The generation of new small compounds reduces the possibility of interference from isotopologs and eliminates the large isotopic mass profiles found in larger molecules. The isotope ratios measured in CRIMS can easily be used to identify and quantify the presence of isotope tags. A chromatogram showing only enriched species can therefore be produced. CRIMS has the important advantage that stable isotopes may be used as tags. This eliminates the use of radioactive material and greatly expands the possible applications of isotopic labeling. These applications can be found in pharmacology, geology, and environmental science. The limit of detection for radio labeled material is better with GC-CRIMS than by on-line radio-chromatography.
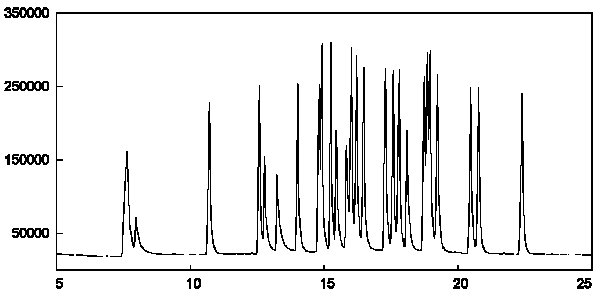
Figure 2a - Chromatogram of qualitative fatty acid methyl ester standard mix as determined by GC-CRIMS analysis. Each component of the standard wa present at an approximate amount of 40 ng.
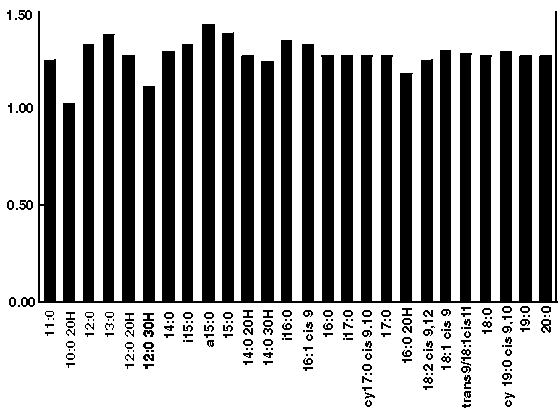
Figure 2b - 13C abundance in qualitative fatty acied methyl ester standard mix as determined by GC-CRIMS analysis. Each of the 26 components of the standard were present at an approximate amount of 40 ng. Average 13C abundance was 1.28 +/- 0.08%
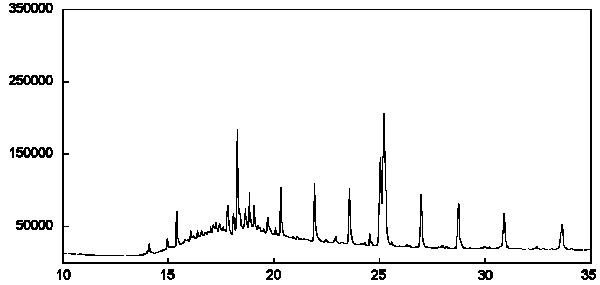
Figure 2c - Chromatogram of fatty acid methyl esters extracted from a pure bacterial culture as determined by GC-CRIMS analysis. 13C abundance ranged from 1.07 to 1.28% with a mean of 1.18%
GC-CRIMS METHODOLOGY
| INSTRUMENTATION | |
| HP
5890 Series II gas chromatograph
HP 5971 mass spectrometer Opthos Instruments microwave power source DB-5 Capillary column, 30 m x 0.25 mm x 0.5 µm film thickness |
|
| GC OPERATING PARAMETERS | |
| Injection | Manual, splitless |
| Sample volume | 1-2 µL in pentane |
| Inlet temperature | 250°C |
| Oven temperature | 100°C for 5 min, 10°C/min to 200°C, 4°C/min to 250°C, hold for 10 min |
| Transfer line temperature | 280°C |
| Carrier gas | Ultrahigh purity He at constant pressure of 16.2 psi (45.4 cm/s) |
| Reactant gas | Anhydrous sulfur dioxide |
| Microwave energy | 70 W forward power and <20 W reflected power |
| MS OPERATING PARAMETERS | |
| Mode | Single ion monitoring at 44, 45 and 76 amu (dwell of 500, 500 and 50 ms, respectively) |
| Solvent delay | 5 min |
| MS temperature | ~175°C |
| Vacuum | <8 x 10E-5 torr |
| MS electrical potential | 2200 eMV |
| DC polarity | Positive |
Conclusions
-GC-CRIMS is suitable for examining bacterial fatty acid methyl esters, yielding acceptable resolution for peak identification.
-Bacterial fatty acid methyl esters in a standard mixture have an abundance of 13C which is only slightly elevated above reported natural abundance levels of 13C.
-Extractions of membrane fatty acids from a pure bacterial culture yielded sufficient material for GC-CRIMS analysis, and 13C abundance was slightly lower than that observed in the qualitative standard.
Future Work
-Determination of isotopic enrichment in bacterial fatty acids extracted from bacteria grown on 13C-enriched substrates.
-Laboratory studies to monitor transport of isotopically labeled bacteria in saturated sediment cores.
-Field studies to evaluate transport of isotopically labeled bacteria in groundwater.
Selected References
CRIMS: chemical reaction interface mass spectrometry, 1994, Abramson, F. Mass Spectrom. Rev., 13:341-56.
Implementation of the chemical reaction interface mass spectrometry technique on a Hewlett-Packard mass-selective detector. 1994. Song, H., Kusmierz, J., Abramson, F., McLean, M. Am. Soc. Mass Spectrom. 5:765-71.
Improved measurement of stable isotope ratios in gas chromatography/mass spectrometry using the microwave-powered chemical reaction interface for mass spectrometry. 1993. Kusmierz, J., Abramson, F. Biol. Mass Spectrom. 22:537-43.
Selective detection of carbon-13, nitrogen-15, and deuterium Labeled metabolites by capillary gas chromatography-chemical reaction interface-mass spectrometry. 1989. Chace, D., Abramson, F. Anal. Chem. 61:2724-30.
Applications of the reaction interface-mass spectrometer technique to the analysis to selected elements and nuclides from sub-microgram quantities of biological macromolecules and xenobiotics. 1988. Chase, D., Abramson, F. J. Res. Natl. Bur. Stand. (U. S.). 93:419.
Accuracy, reproducibility, and interpretation of fatty acid methyl ester profiles of model bacterial communities. 1994. Haack S., Garchow H., Odelson D., Forney L., Klug M. Appl. Environ. Microbiol. 60:2483-93.
Fractionation of fatty acids derived from soil lipids by solid phase extraction and their quantitative analysis by GC-MS. 1993. Zelles L., Bai Q. Soil Biol. Biochem. 25:495-507.
Preliminary observations on bacterial tranport in a coastal plain aquifer. 1997. DeFlaun, M., Murray, C., Holben, W., Scheibe, T., Mills, A., Ginn, T., Griffin, T., Majer, E., Wilson, J. FEMS Microbiol. Rev. 20:473-87.
Development of an adhesion assay and characterization of an adhesion-deficient mutant of Pseudomonas fluorescens. 1990. DeFlaun. M., Tanzer, A., McAteer, A., Marshall, B., Levy, S. Appl. Environ. Microbiol. 56:112-19.
Alterations in adhesion, transport, and membrane characteristics in an adhesion-deficient pseudomonad. DeFlaun, M., Oppenheimer, S., Streger, S., Condee, C., Fletcher, M. Appl. Environ. Microbiol. Submitted.

