- ▶
- Heaters/Source
- ▶
- Agilent Heaters and SensorsMass Spectrometry, Scientific Supplies & ManufacturingScientific Instrument Services 5973 Source Heater Tamper Resistant Allen Wrench 5973/5975 Quad Sensor 5985 Source Heater Assembly Agilent Interface Heater Assembly 5971 Interface Heater

- ▶
- LiteratureApplication Notes Adsorbent Resins Guide Mass Spec Tips SDS Sheets FAQ MS Calibration Compound Spectra Manuals MS Links/Labs/ Organizations MS Online Tools Flyers on Products/Services Scientific Supplies Catalog About Us NextAdvance Bullet Blender® Homogenizer Protocols Micro-Mesh® Literature Instrumentation Literature Agilent GC/MS Literature SIS News / E-Mail Newsletter NIST MS Database - Update Notifications

- ▶
- ManualsNIST 14 Mass Spectral Library Cryo-Trap Model 951 (CO2) Cryo-Trap Model 961 (LN2) Micro Cryo-Trap Model 971 (CO2) Micro Cryo-Trap Model 981 (LN2) SIS Thermal Desorption System Model TD-5 SIS Thermal Desorption System Model TD-4 (old version) SIS Thermal Desorption Software SIS AutoDesorb Model 2000 Operation SIS AutoDesorb Model 2000 Install SIS Desorption Tube Conditioning Oven Direct Exposure Probe Model PC3 High Temperature Direct Probe Model PC-2 SIS AutoProbe Model 2000 SIS AutoProbe Model 1000 (old version) KDS Model 100 Series Syringe Pump KDS Model 310 Series Syringe Pump

- ▶
- NIST 14 Mass Spectral Library (This Page)
NIST Standard Reference Database 1A
NIST/EPA/NIH Mass Spectral Library (NIST 14) and
NIST Mass Spectral Search Program (Version 2.2) June 2014
For Use with Microsoft® Windows
User’s Guide
The NIST Mass Spectrometry Data Center
Stephen E. Stein, Director
Evaluators:
Anzor Mikaia, Principal
Edward White V (EI)
Vladimir Zaikin (EI)
Damo Zhu (EI)
O. David Sparkman (EI)
Pedatsur Neta (MS/MS)
Igor Zenkevich (RI)
Programmers:
Peter Linstrom
Yuri Mirokhin
Dmitrii Tchekhovskoi
Xiaoyu Yang
Documentation and Help:
W. Gary Mallard
O. David Sparkman
U.S. Department of Commerce
National Institute of Standards and Technology
Standard Reference Data Program
Gaithersburg, MD 20899


NIST
14 is available from Scientific Instrument Services, Ringoes, NJ.
The latest details and upgrades on NIST 14 are on our web site:
|
The National Institute of Standards and Technology (NIST) uses its best efforts to deliver a high-quality copy of the Database and to verify that the data contained therein have been selected on the basis of sound scientific judgment. However, NIST makes no warranties to that effect; and NIST shall not be liable for any damage that may result from errors or omissions in the Database.
© 1987, 1988, 1990, 1992, 1994, 1995, 1996, 1997, 1998, 1999, 2000, 2001, 2002, 2005, 2008, 2011, 2014 copyright by the U.S. Secretary of Commerce on behalf of the United States of America. All Rights Reserved.
No part of this Database may be reproduced, stored in a retrieval system, or transmitted, in any form or by any means, electronic, mechanical, photocopying, recording, or otherwise, without the prior written permission of the distributor. Portions of this program are also copyrighted by Microsoft Corp. and © 1984–1996 FairCom Corporation.

.
Microsoft is a registered trademark of Microsoft Corporation, and Windows is a trademark of Microsoft Corporation. ISIS is a trademark of MDL Information Systems, Inc. "FairCom" and "c-tree Plus" are trademarks of FairCom Corporation and are registered in the United States and other countries. All Rights Reserved.
Certain trade names and company products are mentioned in the text to specify adequately the computer products and equipment needed to use this software. In no case does such identification imply endorsement by the National Institute of Standards and Technology of these computer products and equipment, nor does it imply that the products are necessarily the best available for the purpose.
Features in NIST 14 Library and the MS Search Program
Version 2.2 of the MS Search Program
GC Methods / Retention Indices Library
Software Changes from earlier versions of the Search Program
Version 2.0g of the MS Search Program
Version 2.0f of the MS Search Program
Title Bar, Tool Bar, and Status Bar
Spectral Analysis Utilities for NIST 14
Automated Mass Spectrometry Deconvolution and Identification System (AMDIS)
Library Spectrum and Structure Searches
The Search Menu (displayed by Selecting Search on the Menu Bar)
Basis for Interpretation of the Library Search Results
Use with Instrument Data Systems
Use with Third-Party Drawing Programs
NIST Text Format of Individual Spectra
Import and Export of Retention Indices with a Spectrum
RI Export with a Spectrum in a MSP or JCAMP file
RI Import with a Spectrum in a MSP or JCAMP file
APPENDIX 1: Creating AUTOIMP Files Used in NIST MS Search
APPENDIX 2: Using the NIST MS Search Program with ChemStation
APPENDIX 3: Copy ChemStation and Other Database to NIST User Library Format
APPENDIX 4: Using the NIST MS Search Program with Thermo Fisher Corporation Xcalibur Software
APPENDIX 6: Syntax for Some Constraints in Searches
Syntax of Name Fragments Constraint
Syntax of Tags in Comment Constraint
Tags in Comment Constraint for MS/MS Text Information
MS/MS Instrument Type Constraint
APPENDIX 7: Contributing EI Mass Spectra to the NIST/EPA/NIH Mass Spectral Library
Software Registration
Please register your NIST MS product to be alerted of major updates/upgrades. Typically major NIST MS upgrades occur every few years.
(see registration address on your CD cover)
Installation
Windows (NT4/2000/XP/Vista/7/8/8.1)
· Select the Start button on the lower left of the screen. Then select Run from the Start menu. (under Windows 8 or 8.1, open Windows Explorer and run setup.exe from the CD)
· Assuming the CD-ROM drive is D, type D:\SETUP.EXE and select OK. If the CD-ROM drive is a device other than D, replace D with the appropriate drive letter.
· Follow the instructions presented by the SETUP program.
In upgrading from a previous version of the NIST MS Library, there is a possibility to keep both versions.[1] The Installer may behave differently under Windows Vista, 7, 8, 8.1 due to User Account Control security features.
· Note: AMDIS, MS Interpreter, and Lib2NIST are not compatible with Windows NT 4.0.
OS Compatibilities
NIST 14 and MS Search are fully compatible with Windows 2000, XP, Vista, Windows 7 (XP, Vista, and Windows 7 in both 32- and 64-bit editions) and Windows 8.
NIST 14 and MS Search are NOT compatible with Windows 3.x, 95, 98, Me, NT 3.x
NIST 14 and MS Search are partially compatible with Windows NT 4.0. Only NIST MS Search, which provides access to NIST 14 MS Library, will work. NIST 14 version of AMDIS, MS Interpreter, and Lib2NIST are not compatible with Windows NT 4.0. Also needed are:
· Adobe PDF reader to access the NIST 14 Manual
· WinNT4Sp3+MSIE4.0/4.01/4.01SP1+hhupd.exe or WinNT4Sp6+MSIE4.01SP2 or later version of IE to access NIST MS Search html Help
Windows NT 4.0 SP1 and WinNT4Sp3+MSIE4.01Sp1+hhupd.exe under MS Virtual PC 2007 are the earliest versions of Windows that NIST 14 and MS Search have been tested with.
The latest version of AMDIS known to be compatible with Windows NT 4.0 SP1 and MS Search as distributed with NIST 14 is version 2.64 (build Jan. 13, 2006).
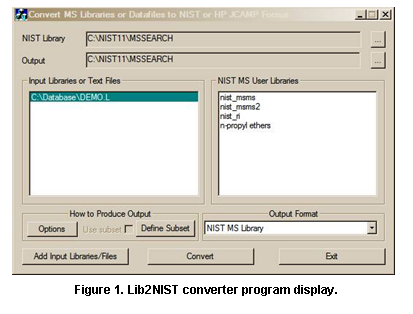
Agilent ChemStation/MassHunter (.L) Format Installation
The following information only pertains to users of Agilent ChemStation/MassHunter.
Agilent ChemStation DataAnalysis (or MassHunter) can send spectra to the NIST MS Search software for library searching. If you purchased the special Agilent format of the NIST EI database (“NIST14.L” folder), you alternately can have ChemStation search the NIST database itself using the Agilent “PBM search” routine rather than using NIST MS Search. You can use either or both methods, whichever you prefer.
If you have this Agilent format, you should manually copy the “NIST14.L” folder from the CD to your hard drive and tell ChemStation DataAnalysis to use it, as described below. The NIST14.L folder MUST be copied from the separate Agilent format installation CD ROM to the \DATABASE folder on the computer that has the ChemStation or MassHunter software. This library will be accessed in the normal way specified by the Agilent software.
(1) Setting Default Mass Spectral Database in ChemStation: In the ChemStation Data Analysis program, select the Spectrum > Select Library menu. In the first row of the Library Search Parameters dialog box, click Browse and select the C:\DATABASE\NIST14.L folder (by default it is was initially C:\DATABASE\DEMO.L) and Click OK. Also, the Spectrum > Set Default Search Engine menu can be used to select whether to use the NIST MS Search or Agilent PBM search by default. This will set the NIST MS library as the default for this session. To have this library remain as the default for future sessions, load the DEFAULT.M method, make the above library change and save it (Method > Save Method menu), and the NIST MS will be the default library for ChemStation searches since other methods are created from this DEFAULT.M method.
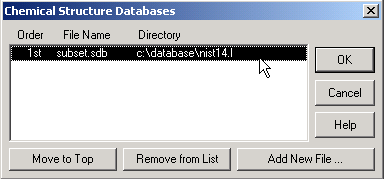
(2) Setting Default Chemical Structures Database in ChemStation: After the default MS library has been set, select the View > Parametric Retrieval menu and click Cancel when the Search Parameters dialog box displays. Then select the Structures > Select Structure Database menu to display the Chemical Structures Databases dialog box. Click Add New File and select subset.sdb in the \Database\NIST14.L folder. If any other older databases are listed (like demo.sdb or older NIST databases like molstru.sdb or subset11.sdb), you probably want to select each and click Remove from List. Click OK to close and finally select the View > Return to Data Analysis menu.
![]() Additional ChemStation
notes are in Appendices 2 and 3.
Additional ChemStation
notes are in Appendices 2 and 3.![]()
![]()
![]()
![]()
![]()
![]()
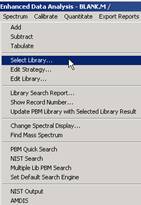
Figure: Selecting the MS library and default search engine in ChemStation.
|
|
|
![]()
![]()
![]()
![]()
![]()
![]() Figure:
Selecting the chemical structure database in ChemStation.
Figure:
Selecting the chemical structure database in ChemStation.
Features in NIST 14 Library and the MS Search Program
NIST 14
The 2014 version of the NIST/EPA/NIH Mass Spectral Library (NIST 14) contains four mass spectral libraries and a Library of retention index (RI)/GC Method. There are two EI Libraries, mainlib and replib.The number of compounds with GC Methods/RI Library in the EI MS Library (mainlib) is 56,216.
|
Contents of the NIST/EPA/NIH (NIST 14) Mass Spectral Library |
||
|
Library Name |
Library Description |
Library Contents |
|
mainlib |
Main EI MS Library |
242,466 spectra |
|
replib |
Replicate spectra Library |
33,782 spectra |
|
nist_msms |
MS/MS Library of small molecules |
193,119 MS/MS spectra for 41,752 ions |
|
nist_msms2 |
MS/MS Library of commercial peptides |
41,165 MS/MS spectra for 3,546 ions |
|
nist_ri |
Retention Index Library |
387,463 RI values for 82,868 compounds |
Version 2.2 of the MS Search Program
Whereas the basic software has not changed since version 2.0g distributed with NIST 11, there have been a number of important features added. These are summarized below.
Features Relevant to Electron Ionization Mass Spectra (Low Resolution)
· Use of Retention Index in library searching: Deviation of experimental RI from the library value optionally used to penalize the Match Factor.
· Older user libraries with RI values can be indexed to allow the RI values to be used in library searching
· Display RI values in a hit list column
· Import, export, and edit compound RI value(s)
· New definitions of derivatives are used for finding replicate spectra
· Optionally display derivative precursor in the Lib. Search hit list
Features Relevant to All Mass Spectra
· All small molecule mass spectra in NIST libraries have InChIKeys linked to PubChem (NIH). These InChIKeys may also be used for searching compound information on the Web or for exact structure searches within NIST libraries.
- Set Prepend/Overwrite/Ask automatic mass spectrum import option.
· Import and display Glycan structures in KEGG Chemical Function (KCF) format and add them to user library mass spectra
Features Relevant to Tandem and High Resolution Mass Spectra
· Display additional MS/MS information in hit lists
· Edit accurate peak m/z, intensities and peak annotations in the built-in mass spectrum editor
· In-source tandem spectra or EI with accurate m/z spectra - High resolution mass spectra
o Import of ‘in-source/EI with accurate m/z’ spectra, which do not include a defined m/z of the precursor ion. To import these spectra, select "In-source /EI (accurate m/z)" in Spectrum Import Options.
o In-source/EI with accurate m/z spectra have accurate ion peak m/z and intensities. The accuracy is set in Spectrum Import Options.
o In-source/EI with accurate m/z spectra may be searched in In-source/EI with accurate m/z and/or MS/MS library with the new In-source HiRes search (ion peak m/z tolerance down to 0.015 ppm or 6x10-5 m/z units), Similarity Simple, Identity Normal or MS/MS Presearch OFF search options. For this purpose, adding Reverse Search option may useful.
o In-source/EI with accurate m/z spectra may be added to a user library. Currently, in-source/EI with accurate m/z spectra added with NIST MS Search to a user library may be searched with In-source HiRes search only with Presearch OFF option. Rebuild the library with Lib2NIST to get in-source HiRes search benefits[2].
· A new MS/MS search for small molecules
o Improved ability to identify compounds with a few dominant peaks – a common situation for Tandem MS.
o More robust scoring for spectra acquired on imperfectly-tuned instruments
· New Mass Spectrum Import Options
o Control the number of decimal places in the input m/z
o Set peak intensity threshold
o Import in-source or EI spectrum with accurate ion peak m/z and intensities.
o Import MS/MS and in-source/EI with accurate m/z spectra from JCAMP files (October 2012, nistms32.dll version 2.1.5.3 or later)
GC Methods / Retention Indices Library
The GC Methods/Retention Indices library has been extended by including summary statistical analysis of all data in the record. This information will be displayed in the spectrum text information even if the Retention Indices box is unchecked in the properties box. The summary data are given in terms of the type of column (standard non-polar(DB-1), semi-standard non-polar(DB-5), or polar(carbowax)), the median value of the retention data for that column type, the deviation and the number of experimental determinations. Note the deviation is calculated as the median absolute deviation. In the example below (for 6,6-Dimethyl-2-methylenebicyclo[3.1.1]heptan-3-ol or pinocarveol ), there are 13 values for the DB5 series of data, 9 for the DB 1 and 12 for the polar column with median absolute deviaions of 4,7 and 8 respectively.
Experimental RI median±deviation (#data)
Semi-standard non-polar: 1138±4 (13)
Standard non-polar: 1135±7 (9)
Polar: 1654±8 (12)
The data will also optionally be shown in the hit list, and the column that will be shown is user selectable.
The Retention Indices checkbox controls the display of the detailed experimental data. The number of GC Methods/Retention Indices displayed is controlled by selecting All or First and entering the number of values to be displayed. The default is to display the first two values. Displaying all values will cause the display to be slow if there are a very large number of literature values.
In addition, for many compounds, an Estimated Kovats Retention Index will be displayed based on the structure associated with the spectrum. These values are reported in index units (iu) with a confidence interval; e.g., “Confidence interval (Ketones): 57(50%) 246(95%) iu” reported for acetone. This means that there is a 50% probability the true value falls within 57 iu of the estimated value and 95% of the time the true value falls within 246 iu of the estimated value. The analysis is based on error measurements of all the data for the class of the compound given; i.e., in this example, ketones. These estimates only display if Retention Indices has been checked in the properties box. The estimates are not displayed in hit lists. If spectra in a user library have associated structures, an Estimated Kovats RI may also be listed for these spectra if the calculation is possible.
The NIST MS/MS Library
MS/MS spectra, along with an increased number of fields relevant to MS/MS data, are provided in a set of files separate from the Main and Replicates data files that comprise the Library of EI spectra. The MS/MS spectra have been provided by contributors, measured at NIST, and extracted from the literature. The preparation of the MS/MS Library revealed that at sufficiently high signal-to-noise measurement conditions, modern instruments are capable of providing very reproducible “library-searchable” spectra. Although collision energy is an important variable in Q-tof and related instruments, spectra vary in an understandable way depending on the compound and instrument type as well as collision-energy conditions. Energy-dependent variation is accounted for by including spectra for most compounds over a wide range of collision energies, ranging from slight to complete dissociation of the precursor ion.
The MS/MS spectra in NIST 14 are provided in two libraries, nist_msms and nist_msms2. The nist_msms library is described in detail below. The second MS/MS library, nist_msms2, contains spectra of compounds that do not have chemical structures displayed. They mostly are commercially available peptides. Spectra from the nist_msms2 library are not displayed in the MSMS tab. Otherwise, the format of the two libraries is similar.
The MSMS tab allows access to the spectra in the nist_msms library. The first time this tab is selected, the three panes of the display are blank with the exception of a single entry in the ion list pane, which is on the left of the tab’s display. This entry is the MS/MS Library. There is a box on the left that has a plus sign . Placing the Mouse pointer on this box and clicking expands to the first two levels (Positive Ions and Negative Ions). There are plus boxes next to both the Positive Ion and the Negative Ion. Clicking on one of these plus boxes results in the display of the next level. This is a list of molecular weights of analytes for which a spectrum was obtained by MS/MS. Clicking on the plus box next to one of these molecular weight listings results in the display of a list of elemental compositions that will also have plus boxes next to them. Clicking on one of the elemental composition plus boxes will result in a display of a list of the compound names that have that elemental composition. In most cases, this is only a single compound. Clicking on the plus box next to the compound name will result in the display of a list of the spectra acquired under MS/MS conditions for this compound. If more than one spectrum was acquired, there is a separate entry for each spectrum. Clicking on the spectrum entry will result in displaying the spectrum information.
The listing of each spectrum begins with MS2.
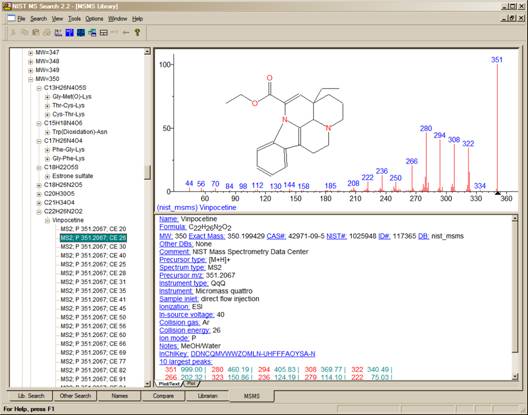
Figure 1. MSMS tab for data acquired using “triple quad”.
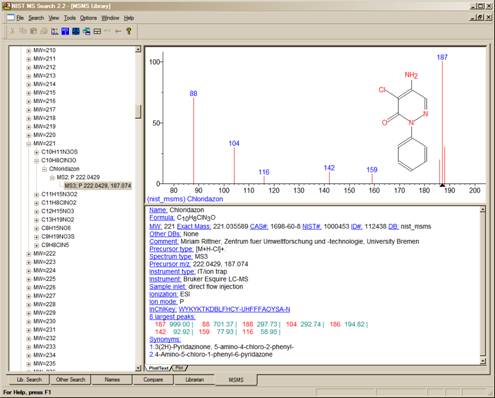
Figure 2. MSMS tab for data acquired using “ion trap MS3”.
For beam-type instruments (e.g., triple quadrupoles, HCD, and Q-tof instruments), the MS2 designation refers to spectra generated by dissociation in a collision cell of a selected precursor ion. For ion trap instruments, the MS2 designation refers to dissociation in the trap after following the isolation of the precursor ion.
In the case of data acquired using an ion trap mass spectrometer, the MS2 may have a plus box next to it. Clicking on this plus box results in a subsequent spectrum listed as MS3, which may also have a plus box. Selection of the plus box next to the MS3 spectrum can result in an MS4 spectrum, which is obtained by the isolation of a product ion as a precursor ion for a subsequent iteration of MS/MS.
Clicking on the MS3 results in the display of the spectrum as seen in Figure 2. The MS2 designation is followed with the precursor ion m/z value and the collision energy for beam-type instruments (Figure 1).
As can be seen from Figure 1 and Figure 2, there is a great deal more information provided in a text format for the spectra in the nist_msms library.
The MS/MS Library can be searched with any of the Other Searches or the Incremental Name Search. All that is necessary is to include the nist_msms library in the Include Libs column of the Search dialog box for the Other Searches or as the single library to be searched in the Incremental Name Search. The Library Spectrum Search and Structure Similarity Search are also applicable. However, the user must understand that MS/MS spectra in general are different from EI spectra as well as the fact that structures in the MS/MS Library refer to the neutral form of the precursor ions.
If the nist_msms library is not present, the MSMS tab is not displayed.
Software Changes from earlier versions of the Search Program
Version 2.0g of the MS Search Program
Below is a list of features new to the previous version, 2.0g, which was distributed with NIST 11.
· A new Exact Mass Search allows searching for exact mass of a precursor (exact molecular weigh) using its mass or exact m/z of a product ion. It takes into account the presence or absence of an electron, adducts or losses; accepts uncertainty in ppm or millimass units; and allows searching for isotopic or monoisotopic peaks.
· A new Exact Mass Constraint similar to Exact Mass Search.
· Ability to use accurate mass to search exact fragment ion mass values in NIST libraries mainlib, replib, and nist_msms using the Any Peaks Search.
- Display of the exact mass with any spectrum that has an associated chemical formula.
· Manage and search as many as 127 separate MS libraries instead of 16, which was the limit in previous versions.
· Optional display of columns in Hit Lists containing the number of synonyms and the number of Other Databases in which the Hit appears. The Hit List can be sorted according to the values in these columns.
· Full compatibility with most recent NIST Peptide MS/MS Libraries.
· Copy selected lines from Hit List or Spec List into Windows Clipboard as tab-delimited Unicode text; this may be conveniently pasted into Excel.
· Spectra in the MSP text format can be imported from the Windows Clipboard into the Spec List via a Right Mouse button (RMB) menu option.
· Searching for words in mass spectrum Text Information Window.
· Optional exclusion of Homologues from the results of Structure Similarity Search.
· Import spectra from mzXML and mzData files.
· Multiple CAS Registry numbers for a spectrum. A small fraction of spectra in NIST 14 has more than one CASrn. Each CASrn may be used in a CASrn Search.
· The MS Interpreter program now enables high mass accuracy for both molecular ions and product ions.
· An alternative peak matching method to improve the reliability of the score when searching noisy MS/MS spectra.
· Several convenience features.[3]
Version 2.0f of the MS Search Program
Below is a list of features new to the previous version, 2.0f, which was distributed with NIST 08.
· A new spectrum search for MS/MS spectra in MS/MS libraries, including the NIST MS/MS libraries.
· Spectra of stereo isomers, tautomers, and derivatives are displayed as replicate spectra.
· Tags (which can act as Field Names) can be entered into the “Comments” field of a user library along with the Field’s contents.
· Hit List can be sorted alphabetically.
· Five new Constraints have been added: Tags in Comment, Peptide Sequences, Peptide Mobile Protons, Peptide Charge, and Peptide Number of Residues. Tags in Comment Constraint, for example, allows searching for the name of the contributor in the EI Library.
· A search for information in the ”Contributor/Comment” field, which may be particularly useful for certain types of information relevant to MS/MS.
· Flexibility has been added to searches using a “Name Fragment in Constraints”. It is now possible to specify the presence or absence of any arbitrary part or parts of a name.
· The Program does not use or require the installation of special NIST fonts.
Guide to the Program
The Program uses tab displays. The tabs:
Lib.
Search, Other
Search, Names, Compare, Librarian, and MSMS
tabs
which are located at the bottom of the Main Window. Selecting a tab
results in the display of a different Window. All tab displays have moveable
divider bars (splitters), some of which can be re-orientated as well as tabs
associated with various panes (windows) within a view. Selecting Window / Tile
Vertically or Window / Tile Horizontally from the main menu displays all
windows.
Lib. Search tab (Figure 3) is used for all searches based on comparison of spectra or structures to a library.
Other Search tab is used for most non-spectral searches, i.e. CAS number, Molecular Weight, Formula, etc.
Names (Incremental Name Search) tab allows you to type in a name and have the program match the name as you type. This search uses all names and synonyms in the specified library
Compare tab allows for easy comparison between spectra. It is, by default, automatically refreshed with the latest spectral search of the library. You can put your own spectra into also.
Librarian tab keeps a history of the recent spectral searches and also can be used for import, editing and user library construction.
MSMS tab gives you access to all the MS/MS data by molecular weight, formula, compound and precursor ion for both positive and negative ions.
In every window, properties of the
pane may be controlled using the Right Mouse Button (RMB) to bring up a
menu from which the context sensitive properties box may be accessed.
Alternate layouts can be selected using the Change layout button ![]() in
the Tool Bar which is just under the Main Menu. A Tool Tip (the
name of a button) is displayed when the Mouse pointer is placed over a Tool
Bar button. There is also a Tool Bar button to restore the default
layout
in
the Tool Bar which is just under the Main Menu. A Tool Tip (the
name of a button) is displayed when the Mouse pointer is placed over a Tool
Bar button. There is also a Tool Bar button to restore the default
layout ![]() (Set default layout). Alternatively,
Default Layout and Change Layout in the Window menu may be used.
(Set default layout). Alternatively,
Default Layout and Change Layout in the Window menu may be used.
All the tab displays have the same Main Menu. The various menu selections available for each of these tabs are described in more detail throughout the manual. The File menu has Save Configuration and Restore Configuration selections which allows all the various tab display configurations to be saved to a specific file name and then recalled. Multiple users may have different ways of configuring each tab that is best for them. This way, each one can restore a specific set of display options. This does not include data search history. There is no change in the search history or Spec List resulting from a Restore Configuration.
The Search selection on the Main Menu allows for quick selection of non-spectral type search. Depending on the choice the selection will transfer you to the correct tab and bring up the options for that search.
The View option on the Main Menu (top of Figure 3) allows the display of the Tool Bar and/or the Status Bar (bottom portion of Figure 3 containing For Help press F1 and Ident). There are also options to turn off the display of the Compare pane in the Lib. Search and Compare tab views.
Tab displays can be brought into focus by placing the Mouse pointer on the tab name at the bottom of the desktop and clicking the left Mouse button. It is also possible to switch between tab views using the hot keys Ctrl‑Tab and Ctrl-Shift-Tab.
Many of the controls for individual window are similar from tab to tab. These are covered in detail for the Library Search Tab but apply for most of the tabs.
The Lib. Search Tab
The first tab (lower left of the Program’s display), Lib. Search, is used with a search of a spectrum or structure against mass spectral libraries: a user library and/or the NIST 14 Libraries (Figure 3). The spectrum and structure searches can be done against multiple libraries (maximum of 127), and the selected libraries can be different from those selected for the various searches found in the Other Search tab. Spectra can be imported into the Spec List from linked mass spectral data systems software, text files containing one or more spectra in the NIST format, or from the Windows Clipboard. Structures in a MOL file format can be pasted into the Spec List for searching from the Windows Clipboard or an ACSII file.
The top pane on the left side of the Lib. Search tab is Spec List (known as the Clipboard Window in previous versions of the Program). The middle pane is a Hit Histogram, showing the distribution of Hits according to the Match Factor values. This Histogram can be displayed with either a linear or log scale on the y axis. The lower left pane contains the Hit List at the bottom. By default, the columns for the number of synonyms, presence in other databases and retention index are not displayed.
On the right side of the movable vertical divider bar are three separate Windows: Search Spectrum/Spec List Spectrum, top; Compare, middle; and Hit List Spectrum, bottom. There are vertical divider bars associated with Spectrum/Text tabs of the top and bottom panes. By selecting View from the Main Menu Bar, the display of the Compare pane can be deselected, it may also be minimized using the splitter bars. If the Mouse pointer is in the Search Spectrum/Spec List Spectrum Window or the Hit List Spectrum pane, the RMB (Right Mouse Button) menu option Change Splitter Orientation can be used to change the orientation (back and forth between vertical and horizontal) of the divider bar between the plot of and text information for the spectrum. In all cases the RMB can be used to access the properties dialog box for each pane within the window.
The pane on the top left of the Lib. Search tab is used for the Spec List, and the pane on the lower left of this Window is used for the Hit List. Note that both the top pane and the bar graph of match factors can be minimized using the splitter bars to maximize the hit list space. The pane on the left of the Other Search and Names tabs is used for the Hit List, and the pane in this position on the Librarian and MSMS tabs is a list of spectra that can be displayed. It is important to know that the Spec List in the Lib. Search tab and the Spec List in the Librarian tab are the same list. Spectra may be copied to the Spec List, edited, and deleted from it. The Hit Lists are read-only. All of these lists can be displayed as text or as graphic structures by selecting the Names or Structures tab at the bottom of the pane, except for the MSMS tab which does not have this alternate display option. Note, only name, match factor, reverse match factor and probability of correct hit are displayed in the graphic mode.
Each Hit List (except the one in the Names tab) and the Spec List can be sorted alphabetically by clicking on the Name Bar at the top of the pane. The Hit Lists in the Lib. Search tab and the Other Search tab have optionally displayed columns showing the number of synonyms, the number of other databases containing the compound and the retention index. These Hit Lists can also be sorted by these columns by pointing to the column header and clicking the left Mouse button.
To copy lines selected in a Hit List or Spec. List into Windows Clipboard in tab-separated text format, press Ctrl+K keyboard keys.
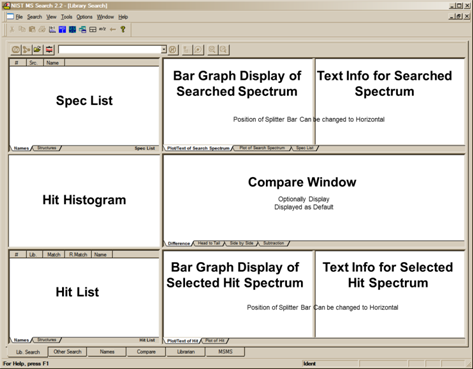
Figure 3. Lib. Search tab.
In some cases, the Hit Lists will also have a plus box in front of the Hit number This symbol means that there are replicate spectra for that Hit. Placing the Mouse pointer on the plus box and clicking the left Mouse button will cause a list of the replicates to appear below the Hit. Some of the names of the replicates will be preceded by R or an M, and some by m or r. If an R or M precedes the name, the spectrum is of the same compound as the Hit and is in the NIST replib Library or the NIST mainlib Library. If the name is preceded by an m or and r it means that the replicate is a derivative or isomer of the Hit, with m being a spectrum in the NIST mainlib and r being a spectrum in the NIST replib.
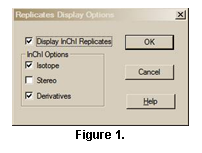 The display of spectra of derivatives
and isomers as replicates of a compound is controlled through the Replicates
Display Options dialog box selected from Options/Replicates using
the Main Menu (Figure
4). Details are found
by clicking on the Help button.
The display of spectra of derivatives
and isomers as replicates of a compound is controlled through the Replicates
Display Options dialog box selected from Options/Replicates using
the Main Menu (Figure
4). Details are found
by clicking on the Help button.
Selecting View/Derivative Parent (Precursor[4]) menu opens Show Derivative Parent for dialog box (Figure 5). Selecting derivative type(s) affects Lib. Search hit list: structures of derivative precursors are displayed instead of derivatives. Only mainlib and replib hits are affected. Deselecting displayed derivative type(s) immediately changes structure display back to derivative. See MS Search help topic "Show Derivative Parent" for more details.
The Spectral Plot presentation pane (upper right of the Window in the default display) of all tab views except the Compare tab, have settable attributes that are found in the Properties dialog box described below. By default, the Plot pane displays the primary name of the spectrum in the lower left hand corner of the pane. In addition the library from which the spectrum was drawn precedes the name in parenthesis. Optionally, the compound’s Molecular Weight (nominal mass), CAS registry number, and Formula (elemental composition) can be displayed. This legend display can be forced to wrap so that all characters will be visible (Figure 6). Also optional, a structure (if associated with the spectrum) can be displayed on the spectrum. If a structure has been requested and one is not assorted with the spectrum, a colored ? will appear. Labeling of the peaks is either by default or the Plot tab of the Properties dialog box has a “Label peaks greater than (%)” option followed by a text entry box allowing for the input of a number. If the spectrum is to be displayed without peak labeling, enter 100 in this box or deselect m/z Label and Text Label options. It is also possible to use this Properties dialog box to change the colors of the mass spectral peaks, scale, labels, bonds and rings in structures, atomic symbols in structures, and the spectrum’s background. Selecting the Set default layout button on the Tool Bar will restore colors to the original values.
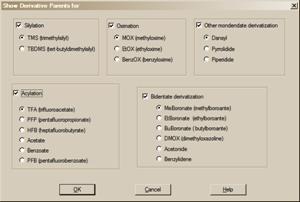
Figure 5. Select anyone of the five Derivative type check boxes; then select the subcategory radio button.
The Text Info pane, by default, is to the right of the Spectral Plot presentation in the Lib. Search tab. This orientation can be changed so that the Text Info pane is just below the Plot presentation by using the RMB menu selection Change Splitter Orientation. In the other tab views where both the plot of the spectrum and the text information are displayed, these two panes are oriented by default with the Plot pane above the Text Info pane. Again, this orientation can be changed using the appropriate selection on the RMB menu when the Mouse pointer is on either of the two panes. The Hit Text Info tab (Text Info tab in some tab views) of the Properties dialog box has a number of selectable display options in the form of check boxes (Compound Information, m/z intensity list, Ten Largest Peaks, Synonyms, and Retention Indices). The Compound Information is: the primary name associated with the spectrum in its source library; the Formula (elemental composition); MW (nominal mass); Exact Mass (if there is an elemental composition associated with the spectrum); CAS registry number; NIST Number (sequential number assigned to spectra in the NIST EI Archive); ID number (position in the library containing the spectrum); name of the library containing the spectrum; names of Other databases in which the compound can be found, if any (displayed only with mainlib and replib spectra); Contributor (NIST Library)/Comments (user libraries); and Related CAS registry numbers, if any (NIST DB only). The other check boxes of these first four selections are self-explanatory.
If Retention Indices is checked and there are records containing literature GC methods and Retention Indices, these records will be displayed following the display of synonyms (if selected). As pointed out earlier, by default, only the first two records are displayed. If there are more than two records, <…> will appear just below the last line of the bottom record. The Properties dialog box allows the setting for the display of a larger selected number of records or all of the records. The record is searchable using the Find selection on the RMB menu when displayed in a Text Info pane. All text is searched so column types, active phases, authors or text in the article titles can be searched. Only the records displayed are searched (Figure 7).
Figure 6. The Spectral Plot display of a spectrum in the Name tab with the nominal mass, CAS registry number, elemental composition name of the library containing the spectrum, and the name associated with the spectrum. The display is wrapped, the structure is shown, and the peaks are not labeled.
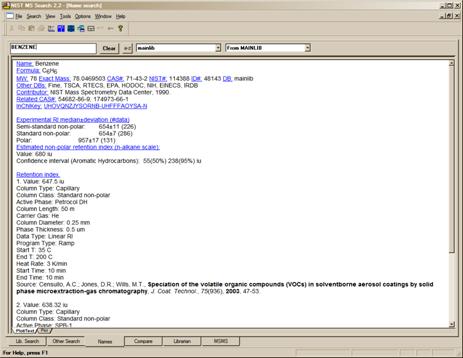
Figure 7. The Text Info display with Compound Information and Retention Indices as the only selected display options.
It should be noted that the GC Methods/RI Library is also available separate from NIST 14 with its own Search program, which allows for more extensive use of this Library.
 The RMB menu selection Properties
will result in the display of a multiple-tab dialog box (Figure 8). The tabs at the top are
self-explanatory. This allows customization of the appearance of the display
and control of what is displayed such as a structure on a spectrum, the 10 most
intense peaks in the spectrum in the text display, etc. Colors of structures,
mass spectral peaks and axes, and fonts used for alphanumeric characters can
also be set. Each pane can have its own customized set of attributes. This
customized display can be saved using the File/Save Configuration
command from the Main Menu Bar. Different saved configurations can be
recalled by using the File/Restore Configuration command from the
Main Menu Bar. Separate customizations can be established for each of
the six separate tabs. This includes selected libraries to be searched.
The RMB menu selection Properties
will result in the display of a multiple-tab dialog box (Figure 8). The tabs at the top are
self-explanatory. This allows customization of the appearance of the display
and control of what is displayed such as a structure on a spectrum, the 10 most
intense peaks in the spectrum in the text display, etc. Colors of structures,
mass spectral peaks and axes, and fonts used for alphanumeric characters can
also be set. Each pane can have its own customized set of attributes. This
customized display can be saved using the File/Save Configuration
command from the Main Menu Bar. Different saved configurations can be
recalled by using the File/Restore Configuration command from the
Main Menu Bar. Separate customizations can be established for each of
the six separate tabs. This includes selected libraries to be searched.
At the top left of the Lib. Search tab view (Figure 3) is the Button Bar. The meaning of each of these buttons is shown below:
![]()
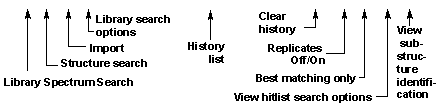
Figure 9. Lib. Search’s Button Bar.
The Go button will launch a search of the highlighted spectrum in the Spec List. The Structure Search button will perform a structure similarity search of the highlighted structure in the Spec List. The Import button (with the file folder icon) is used to import spectra from a text file. The Library Search Options button is used to display the dialog box used in selecting libraries to be searched, search types, and other factors associated with the search of spectra and structures.
The drop‑down list box contains the Search History. A previous search’s display can be restored by highlighting an entry in this list. NOTE: the Program can be set to automatically clear the Hit List and/or the Spec List by selecting the Clear History on Exit check box in the Hit List tab and/or Plot/Text of the Spec List tab in the Library Search Properties dialog box displayed by selecting Properties from the RMB menu.
The Clear History button is used to clear the search history. The Replicates On/Off button will toggle the listing of replicate spectra on and off when the mainlib Library of the NIST/EPA/NIH Library is searched.
The Best matching only button will force the hit list to display only the hit with the highest match factor for any given CASrn. This is the. This feature is especially useful when more than one library is being searched, and some of the spectra in these libraries have the same CASrn’s associated with them. In the case of an MS/MS Search, the best matching spectrum is selected out of Hits that have the same CASrn or names and charge.
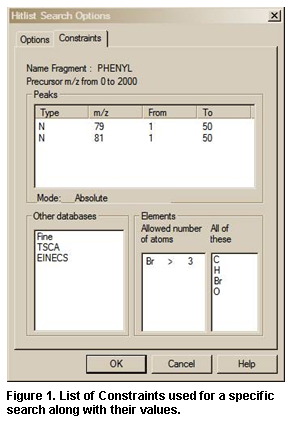 The View hit list search options
button will result in the display of a dialog box with two tabs if constraints
have been used in the search. The Search or Options tab
(depending on whether the Hit List Search Options are for a Lib.
Search or an Other Search carried out) lists the libraries searched
(and type of Search and Search Options for Library spectrum Searches
only). The Constraints tab (if present) lists all different constraints
along with values used in the search (Figure 10 and Figure 11).
The View hit list search options
button will result in the display of a dialog box with two tabs if constraints
have been used in the search. The Search or Options tab
(depending on whether the Hit List Search Options are for a Lib.
Search or an Other Search carried out) lists the libraries searched
(and type of Search and Search Options for Library spectrum Searches
only). The Constraints tab (if present) lists all different constraints
along with values used in the search (Figure 10 and Figure 11).
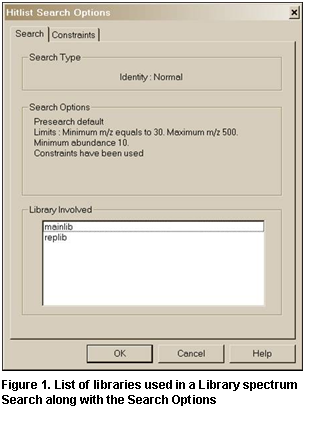 It should be noted that even though a
Library spectrum Search is carried out against a list of libraries (Figure 28) that does not include mainlib
and replib, these two libraries will be listed as having been included (Figure 11) as long as the Display InChI Replicates check
box is selected in the Replicates Display Options dialog box (see Figure 4). This will result in the display
of the symbol indicating replicates were found when the user-only library was
searched. This means that InChI search options are available for those
spectra in libraries like the Wiley Registry of Mass Spectra and various other
libraries provided by Wiley and Others in the NIST format, which have CASrn
present in mainlib.
It should be noted that even though a
Library spectrum Search is carried out against a list of libraries (Figure 28) that does not include mainlib
and replib, these two libraries will be listed as having been included (Figure 11) as long as the Display InChI Replicates check
box is selected in the Replicates Display Options dialog box (see Figure 4). This will result in the display
of the symbol indicating replicates were found when the user-only library was
searched. This means that InChI search options are available for those
spectra in libraries like the Wiley Registry of Mass Spectra and various other
libraries provided by Wiley and Others in the NIST format, which have CASrn
present in mainlib.
Two other Important Points about the use of Constraints:
When a Constrained Search is carried out, the Search is performed as if there was no constraint. Only the spectra that were found in the Search that fall within the constraint criteria appear in the Hit List.
The number of different constraints that are available exceeds the display area; a slider bar on the right side of the Constraints Window in the Search dialog box allows access to all constraints. The Constraint Window will only allow for the display of six constraint options at a time. Once values have been entered for any of these options, those values will remain until individually changed or deleted. Once an available Constraint has been selected (check mark in check box entered by putting the Mouse pointer on the check box and clicking the left Mouse button), the values previously entered for that constraint will be used when a Constrained Search is performed. Because if a Constraint is selected and is not visible in the Window, a bad result may occur, the program displays the number of constraints selected. A Clear All button has been added to the Constraints tab of the Search dialog box. The Clear All button removes the checks in the various constrains, it does not change the values in each of the constraints. It is recommended to click on this button before starting a new Constrained Search and reselecting any desired Constraints. Whenever any Constraint is selected, the Use Constraints check box is automatically selected. Clicking on any individual Constraint’s check box in the Constraints Window causes appearance of the Constraints tab of the Search dialog box to change to allow for the entry (or edit) of values for that Constraint at the same time the Constraint is selected or deselected. The appearance can also be changed to that for a specific constraint by clicking on the constraint description away from the check box.
The last button on the right, the View substructure information button, displays the Substructure Information dialog box. The Substructure Information dialog box contains a list of the probability of the presence and absence of substructures that may be associated with a compound that produced the searched mass spectrum along with additional predictions about this compound’s molecular weight and estimate of the numbers of atoms of chlorine and/or bromine that may be contained. This is especially useful with a Neutral Loss, Hybrid Neutral Loss Similarity, or MS/MS In EI Similarity Search. An example of the use of Substructure Information resulting from an MS/MS in EI Similarity Search of the EI Library is provided later in this manual.
Title Bar, Tool Bar, and Status Bar
At the top of the display of each of the Program’s tab is a Title Bar. The contents of a title bar of the active tab are also displayed in the Title Bar of the Program Window. When the Lib. Search or Other Search tab is displayed, this Title Bar will contain information about the currently displayed search. In the case of a Library spectrum Search, this will include the search type used, whether or not constraints were used, and if any type of limits were applied to the search. In the case of the Other Search tab, the Title Bar will contain the type of search that was done (i.e., Formula, Molecular weight, Any peaks, etc.) and whether or not constraints were used. When the Compare tab is displayed, the Title Bar contains the number of spectra available for comparison regardless of the number displayed.
At the bottom of the Program’s display, below the Program’s tabs, is the optionally displayed Status Bar. The Status Bar’s display can be turned off by selecting or deselecting Status Bar from the View menu. When a Library spectrum Search is being conducted, the left side of the Status Bar will have displayed “Comparing XXX Library spectra with submitted spectrum” where XXX is the number of spectra that were found during the Presearch (see Search Algorithms in Appendix 5). If Off has been selected in the Presearch area of the Search tab of the Library Search Options dialog box, XXX will represent the total number of spectra in the libraries being searched.
There are five fields on the right side of the Status Bar. The left-most of these five fields will have displayed the selected search type in the “Spectrum Search Type” area of the Search tab of the Library Search Options dialog box (Neutral Loss, Hybrid, or Simple if the Similarity Search is selected; and Quick or Ident depending on whether a Quick or Normal Identity Search has been selected).
The next four fields to the right of the box indicating the selected search type pertain to the currently displayed search results. The first box to the right contains the type of search that was used in the displayed search results. The next box to the right will have contained “Rev” if the Hit List is sorted according to the Reverse Match value (the Match Factor obtained by ignoring all peaks that are in the sample spectrum but not the library spectrum). To set this mode, select Reverse Search check box in the “Spectrum Search Options” section of the Search tab of the Library Search Options dialog box. The third box will contain the number used as a precursor MW value in a Neutral Loss or Hybrid Similarity Search. If an Identity Search has been carried out, the fourth box will contain “Penalize” if the Penalize rare compounds check box was selected in the “Spectrum Search Options” section of the Search tab of the Library Search Options dialog box.
The Tool Bar’s display (Figure 12) is optional by selecting it from the View menu. The functionality of its button Switch to Caller and Print Report is also available on the File menu; MS Interpreter, and AMDIS also may be launched from the Tools menu.
Figure 12. NIST MS Search Tool Bar.
A Tool Tip is associated
with each of the buttons with an explanation of the button’s function. The ![]() button
will return all displays on the currently viewed tab to the default values.
The
button
will return all displays on the currently viewed tab to the default values.
The ![]() button toggles through three separate
views for the currently displayed Program’s tab.
button toggles through three separate
views for the currently displayed Program’s tab.
The m/z range button displays a dialog box (Figure 13) to fix the starting and ending m/z values for bar-graph spectra displays, and to choose whether the peaks should be labeled with neutral losses or with their m/z value.
An explanation of each of the items in this dialog box can be found by selecting the dialog box’s Help button. The Anchors dialog box is used when viewing spectra with a neutral loss label. Selecting Anchors will result in the display of the Anchors dialog box, which is explained by selecting the Help button in that dialog box.
The two radio buttons at the top of this dialog box have to do with labeling spectra normally or with natural loss values. Normal display is the most common.
Neutral Loss Display
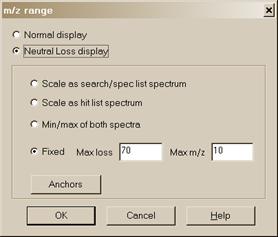
Figure 13. m/z range (for Spectra Display) dialog box.
One of the important tools in deducing a structure from a mass spectrum is to look at the mass spectral peaks as losses from the molecular ion rather than as individual peaks. In this way of viewing the mass spectrum, the peak at m/z 269 in methyl stearate becomes the [M - 29]+ peak (loss of an ethyl radical) and the peak at m/z 267 becomes the [M - 31]+ peak (loss of a methoxy radical). See Figure 14.
The program can display a spectrum with neutral loss labeling with the library spectra and search spectrum be labeled as neutral losses with the first label on the right-most peak being 0 (Figure 15). To activate this display, select Neutral Loss Display from the RMB menu. Selecting Neutral Loss Anchors is explained in the Help files (topic m/z range). NOTE: If you do not know the molecular weight of the search spectrum, the program will attempt to estimate it. This value may be overridden by using the librarian to edit the spectrum and specifying a molecular weight and then using the edited spectrum to run the library search.
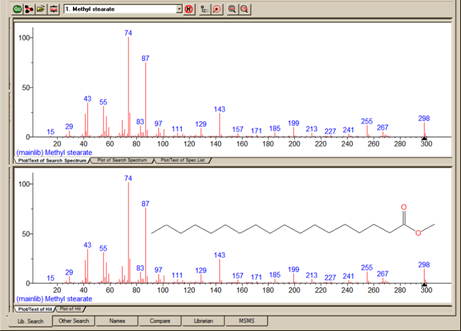
Figure 14. Nominal spectrum labeling where the top spectrum is of an unknown compound and bottom spectrum is of the first Hit from a search of the NIST/EPA/NIH Mass Spectral Library (NIST 14).
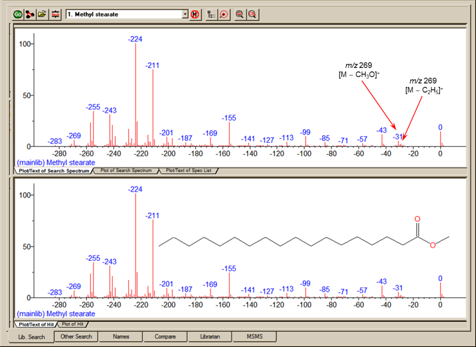
Figure 15. Neutral Loss spectrum labeling where the top spectrum is of an unknown compound and bottom spectrum is of the first Hit from a search of the NIST/EPA/NIH Mass Spectral Library (NIST 14).
The Other Search Tab
The second tab display, Other Search (Figure 16), is used to perform a MW (Nominal Mass), Exact Mass, Formula (elemental composition), Any Peaks, Sequential Method, CAS registry number, NIST Number, or ID Number Search. The NIST Number Search can only be carried out on the NIST Main Library (mainlib) and the Replicates Library (replib). NIST Numbers establish correspondence between spectra in different releases of the NIST/EPA/NIH MS Library. The ID Number Search can only be carried out on one library at a time. The other options in this group can search multiple libraries (maximum of 127), and different sets of libraries can be specified for each different type of search.
As shown in Figure 17, another new feature in v.2.2 of the Program is the display of columns showing not only the number of synonyms, the number of other databases in which the Hit can be found but also the retention index (RI) if available. Not only are these columns available for the Hit List of searches performed in the Other tab, but also in the Hit List displayed in the Lib. Search tab. The display of these columns is controlled by the Hit List tab of the Properties dialog box displayed by selecting the Properties option from the RMB menu displayed when the Mouse pointer is in a Hit List pane.
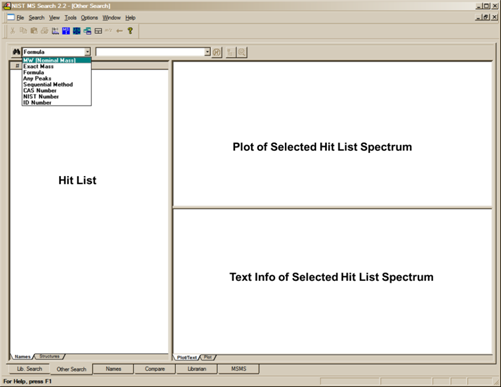
Figure 16. Other Search tab.
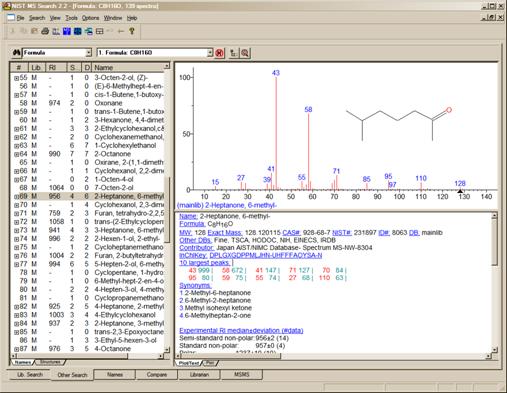
Figure 17. Results of a Formula Search showing the number of “synonyms” for the Hits ,the number of other “databases” in which the Hits can be found and the RI value (if present) columns.
The Names Tab
The third tab, Names, is used for the NIST signature Incremental Name Search (Names) (Figure 18). The Name Search can only be performed against one mass spectral library at a time. The library to be searched is selected by use of the Library to search drop-down list box next to the a–z button. If the a–z button is selected, the only input in the text entry field on far left just below the button bar that will be taken is that of letters a–z. If the a–z button has not been selected, numbers as well as letters can be input. Names containing Greek alphabet characters can be pasted into the text entry field. Typed dashes and other punctuation characters will not be used. All letters are entered as upper case regardless of the input.
NOTE: The drop-down list box just to the right of the a–z button is used to select the library to be searched. This search method allows for the browsing of user libraries.
The second list box only appears when the library selected is one of the NIST mainlib is searched. It is used to display replicate spectra for the NIST 14 mainlib Library. If there are no replicate spectra for the highlighted selection in the Hit List, this list box will not appear on the display. When Replicates are available they will appear in the list box as either Replicate followed by a sequence number or *Replicate. Replicate is a spectrum of the same compound that is in the replib Library. *Replicate, according to Replicates options identifies a spectrum that is of a heavy isotope variant of the compound, a stereoisomer of the compound, or a compound, which has the same derivative precursor.
The Name Search used with the NIST/EPA/NIH Mass Spectral Library uses not only the main name but also any synonyms. As an example, the mainlib compound name for acetylsalicylic acid is the common name Aspirin. This compound has 157 synonyms and its spectrum can be displayed by entering any of these names including trade names like Bayer (Figure 18).
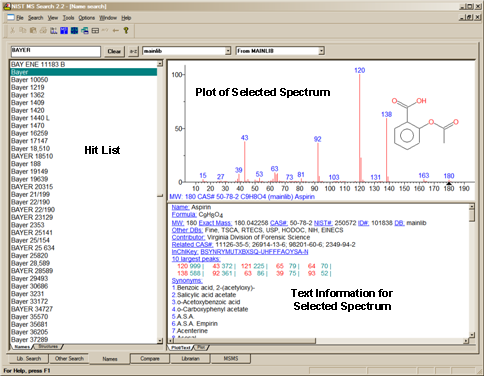
Figure 18. Incremental Name Search tab.
Note, the use of synonyms is also supported in user libraries. Thus if a spectrum is given a synonym “Lab Book 4-PG-22”, then the data can be retrieved by that name using the incremental search.
The Compare Tab
The fourth tab, Compare (Figure 19), allows the display of a sample spectrum and multiple spectra from the Hit List. Other spectra can also be put into the Compare display. A selected spectrum in the lower part of the Compare tab can be displayed as various types of comparisons with the spectrum in the upper part of the display by selection of one of the tabs in the Compare pane (Difference, Head-to-Tail, Side-by-Side, Subtraction).
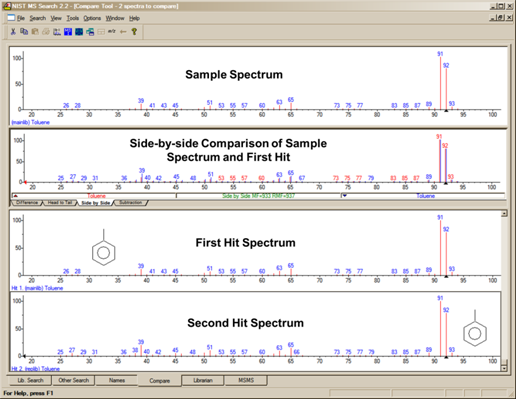
Figure 19. Compare tab.
The Librarian Tab
The fifth tab, Librarian (not shown), is used to deal with spectra that are in user libraries or that are to be added to user libraries, or have been imported or searched. The layout of this tab is the same as the tab used for the Incremental Name Search with the exception that the far left pane contains the Spec List rather than a Hit List. This is the same Spec List as displayed in the upper left pane of the Lib. Search tab. The Librarian tab allows for the editing or creating of spectra. This editing not only allows m/z intensity pairs to be edited, deleted, or added; but also allows CAS registry numbers, elemental compositions, structures, names, synonyms, and structures to be edited, added, or deleted.
When a splitter bar divides a pane, the position can be changed by selecting Change Splitter Orientation on the RMB menu, displayed when the pointer is in the pane.
Zoom. Placing the Mouse pointer on a mass spectrum plot or spectra comparing bar-graph plot, holding the left Mouse button down and dragging the Mouse pointer along a diagonal will result in drawing a zoom rectangle. After releasing the Mouse button, a zoomed spectrum will be displayed; the cursor will change to a magnifying glass. To return to the normal display, select Zoom Out from the RMB menu or double-click.
Additional information on the Librarian and MSMS tabs is provided later in this manual under User Libraries and MS/MS Searching.
Spectral Analysis Utilities for NIST 14
Automated Mass Spectrometry Deconvolution and Identification System (AMDIS)
Included as a separate utility, AMDIS attempts to reconstruct original mass spectra for individual components in arbitrarily complex GC/MS and LC/MS chromatograms; and if a target library is provided, AMDIS can directly identify target compounds. AMDIS is especially useful when a single reconstructed total ion current (RTIC) chromatographic peak represents multiple components. Regardless of each component’s concentration, pure mass spectra are deconvoluted for analyses. AMDIS was developed by NIST under a contract from the Defense Threat Reduction Agency (DTRA), Department of Defense, for verifying of compliance with a major international treaty (Chemical Weapons Convention) as ratified by the United States Senate in 1997. In order to meet the rigorous requirements for this purpose, AMDIS was tested against more than 30,000 GC/MS data files accumulated by the EPA Contract Laboratory Program without a single false-positive for the target set of known chemical warfare agents. Although this level of reliability may not be required for all laboratories, this shows the degree to which the algorithms have been tested. AMDIS can also be used to identify individual non-target components in complex samples. It has been widely used in the flavors and fragrance industry, forensics, the study of natural products, and for the identifications of metabolites.
AMDIS has been designed to reconstruct “pure component” spectra from complex RTIC chromatograms even when components are present at trace levels. For this purpose, observed chromatographic behavior is used along with a range of noise‑reduction methods. AMDIS is distributed with specialized libraries (environmental, flavor and fragrance, and drugs and toxins) that were derived from the NIST 02 MS Library. AMDIS has a range of other features including the ability to search the entire NIST 14 MS Library with any of the spectra extracted from the original data file. It can also employ retention-index windows when identifying target compounds and can make use of internal and external standards maintained in separate libraries. A history list of selected performance standards is also maintained.
The instruments that are supported in their native file format are: †
Agilent ChemStation (GC-MS & LC-MS) and MS Engines
BrukerDALTONICS GC-MS (*.MSF)
Finnigan (GCQ, INCOS, ITDS, ITS-40, ITD-700/800, ITMS)
Inficon (*.acq)
JEOL/Shrader (*.lrp)
Kratos Mach3 (*.run)
Micromass (MassLynx, NT Formats) (RAW)
mzXmL/mzData
Perkin-Elmer TurboMass (RAW) and ITD-700 (*.ms)
Shimadzu MS Files QP5000 and QP5050 (*.R##)
Thermo Fisher Xcalibur (RAW)
Varian (MS, SMS, XMS)
In addition, the NetCDF format, which is supported by many manufacturers, is supported by AMDIS. Other formats will be added in future releases. Contact NIST for future information.
† The use of specific product and trade names does not indicate an endorsement by the National Institute of Standards and Technology. The specific file formats are supported in order to ensure that the data is transferred to AMDIS with no information loss. The specific formats and names are covered by the respective copyrights of the individual companies.
MS Interpreter
This tool was developed to aid NIST evaluators in their analysis of mass spectra. In one integrated program, it enables a variety of calculations on a mass spectrum, using a proposed chemical structure if available. Spectra and structures are associated in the spectral editing component of the Librarian tab of the MS Search Program discussed in USER DATABASES section and APPENDIX 3 of this manual. MS Interpreter is activated for an individual spectrum with the Mouse Pointer on a Plot or Text Information pane or by highlighting the name or structure of a compound in a Hit List and selecting MS Interpreter on the Send To menu of the RMB menu. This action opens MS Interpreter with the spectrum and associated structure (Figure 20). MS Interpreter can also be opened from the Tools menu or a button on the Tool Bar. If a spectrum/structure has been opened in MS Interpreter and then the program closed and then reopened from either the Tools menu or the button on the Toolbar, the last contents will be displayed. If MS Interpreter is started by either of these two actions just after starting the Program, both the Structure pane and the Spectrum pane will be empty. Spectra can be imported from MSP files (text files containing spectra in the NIST MS Search format), JCAMP-DX, and SDfiles. If the folder also contains a *.STB structure file that has the same name as the MSP file, that structure will be associated with a spectrum in MS Interpreter. When a structure/spectrum pair is Saved from the MS Interpreter’s File Menu, two files are created: one containing the spectrum (*.MSP) and the other a structure (*.STB).
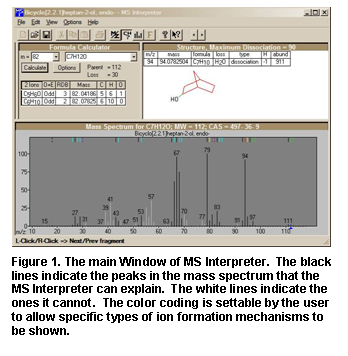
 Peaks in the spectrum originating as
a logical fragment of the molecule (M+· or MH+) are marked at the top of the
spectrum display; and corresponding fragments, derived from thermodynamic
approximation, may be highlighted by clicking on one of these marks. It also
allows for keeping track of important neutral losses, both from the precursor
or a derived ion and to find all possible formulas for any peak or neutral
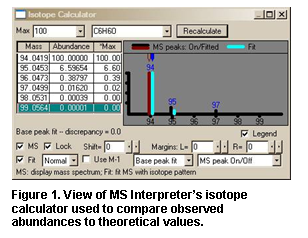
loss. Isotopic patterns (Figure
21) for any cluster can
be separately examined. These isotope clusters can be compared to theoretical
predictions subject to a number of user-specified constraints. MS
Interpreter is a separate program that also has an icon in the NIST Mass
Spectral Database program folder. It also has a simple Help screen that can
be called from the program by selecting Help on its Menu Bar or
by pressing the F1 key. Information about the control currently located under
the mouse pointer and possible user actions is displayed in the Status Bar
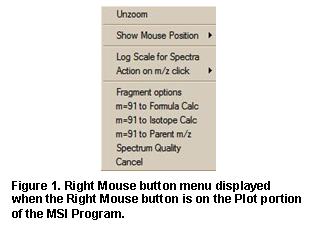
located at the bottom of the program’s main Window. Placing the Mouse Pointer
in the mass spectrum display and clicking the Right Mouse button will result in
a somewhat self-explanatory RMB menu. Before using the program, explore this
RMB menu (Figure 22).
Peaks in the spectrum originating as
a logical fragment of the molecule (M+· or MH+) are marked at the top of the
spectrum display; and corresponding fragments, derived from thermodynamic
approximation, may be highlighted by clicking on one of these marks. It also
allows for keeping track of important neutral losses, both from the precursor
or a derived ion and to find all possible formulas for any peak or neutral
loss. Isotopic patterns (Figure
21) for any cluster can
be separately examined. These isotope clusters can be compared to theoretical
predictions subject to a number of user-specified constraints. MS
Interpreter is a separate program that also has an icon in the NIST Mass
Spectral Database program folder. It also has a simple Help screen that can
be called from the program by selecting Help on its Menu Bar or
by pressing the F1 key. Information about the control currently located under
the mouse pointer and possible user actions is displayed in the Status Bar
located at the bottom of the program’s main Window. Placing the Mouse Pointer
in the mass spectrum display and clicking the Right Mouse button will result in
a somewhat self-explanatory RMB menu. Before using the program, explore this
RMB menu (Figure 22).
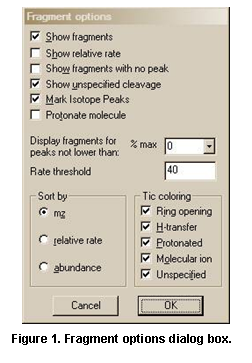
Selected ions on the plot of the mass spectrum can be sent to Formula Calculator and the Isotope Calculator using selections on this menu. A specific m/z value can be locked as the position to display calculated losses from. In addition, the Fragment options dialog box can be displayed (Figure 23).
More detailed information can be obtained about the use of this utility by pressing the F1 key with the Mouse pointer in one of the Program’s panes.
Exact masses of fragment ions
Structure view displays not only nominal m/z but also exact mass of the predicted fragment ion.
Pasting a chemical structure into the Structure view
· Copy a structure from a structure editor (molfile format) into Windows Clipboard;
· In MS Interpreter, click Structure view title bar to activate it;
· Click Paste button or select from the menu: Edit/Paste or press Ctrl-V.
Pasting a mass spectrum or a chemical structure into Mass Spectrum view
Contents of a file containing a mass spectrum or a structure or both (SDfile) may be pasted into the Mass Spectrum view.
Pasting a peak list into the Mass Spectrum view
· Copy a peak list into the Windows Clipboard;
· In MS Interpreter, click Mass Spectrum Window to activate it;
· Click Paste button, or select from the menu: Edit/Paste, or press Ctrl-V.
Notes
- the peak list must be made out of m/z and intensity pairs
separated by any of the following characters:
space , ; : ( ) [ ] } | Tab - each line must contain one or more m/z and intensity pairs;
- if at least one line contains an odd number of values, then the program reads only first two numbers (one peak) from each line; a warning is displayed;
- all peaks must be in order of either ascending or descending m/z;
- all m/z are rounded to the nearest integer by the MS Interpreter;
- there is no need to normalize intensities to 999.
Pasting MSP, SDF, MOL, and JDX (JCAMP-DX) files from Windows Explorer
· MSP, SDF, MOL, and JDX files may be copied from the Windows Explorer and pasted into the Structure or Mass Spectrum view of MS Interpreter.
· The files of the same type may be dragged from the Windows Explorer and dropped into the same MS Interpreter views. In all cases, only the first spectrum and/or structure from the first selected file is/are imported.
· The name of the imported file appears on the MS Interpreter Title Bar and in the MRU (most recently used) list in the File menu.
Copying calculated isotope envelope table from the Isotope Calculator into Windows Clipboard
· Right-click any line in the Isotope Calculator table
· Select Copy.
This will copy into the Windows Clipboard a tab-delimited text, which is suitable for pasting into a MS Excel spreadsheet or text editor. The context menu and the text contain words “Default Isotope Table” or the name of the file from which a custom isotope composition was imported (see the next section.)
Using custom isotope composition table in Isotope Calculator and Formula Calculator
· Open file MS_Interpreter_isotope_table_test_01.itbl in a plain text editor. This file is installed in the same folder as MS Interpreter, typically, c:\nist11\mssearch
· Read the notes in the file
· Edit the file and save it under another name
· From MS Interpreter menu, select File/Import Isotope Table
· In the Open Isotope File dialog box, select the newly created file and click the Open button.
· To restore the original table, import file MS_Interpreter_isotope_table_default.itbl. The original table is restored automatically after both MS Interpreter and MS Search are shut down.
· Every time a *.itbl file is imported, a *.itbl.log file is created or appended. This file contains the date, time, a full pathname of the .itbl file, and a list of differences (if any) between the current and the imported isotope table or a reason for the failure to import the new table.
Example Searches
Library Spectrum and Structure Searches
The Program can be used to find spectra that are similar to that of an unknown compound. This can be accomplished by importing the spectrum from a third-party mass spectral data analysis program or from a text file (*.MSP). A spectrum can also be created using the Spectrum Information dialog box displayed by selecting the New button on the button bar of the Librarian tab to enter the m/z and intensity pairs that make up a mass spectrum. The Library spectrum Search and the Structure Search are usually carried out from the Lib. Search tab by clicking on the Library Search button (Go) or Structure Search button (next to the Go button). Another way of initiating a search is to select a spectrum in any Hit List or the Spec List, Plot pane or Text Info pane in any tab, or in a Compare pane; display the RMB menu and click with the Right Mouse button any spectrum in a List of spectra or Subtraction spectrum in the Compare pane; and select the desired search (Library Search for a spectrum search or Structure Similarity Search for a structure search). To search for more than one spectrum at once, make a multiple selection in any List of spectra (Hit List, Spec List, or Other Search List except Name Search List), display the RMB menu, and select Library Search or Structure Similarity Search. Regardless of the tab display, when one of these searches is carried out, the view will change back to Lib. Search tab. In the default display of the Lib. Search tab, the Hit List is on the lower left. The third column (from the left) of the Hit List contains the Match Factor. Match Factor 1000 for the Structure Search does not mean an exact match; it means that the found structure is a homologue of the submitted structure. Match Factors for the Spectrum Search cannot exceed 999.
Spectra and structures can be imported from files or third-party programs through various types of automations, described later in this section. Spectra or structures can be imported from a file by selecting File from the Main Menu Bar and selecting Open from the File menu (Figure 24). Selecting a file will result in a display of the Structure Import dialog box or the Spectrum Import dialog box. The Structure Import dialog box is self‑explanatory. The Spectrum Import dialog box (Figure 25) is more complex. When a file containing more than one spectrum is selected, one or more selected spectra or all of the spectra can be imported by using either the Import Selected or the Import All button, respectively. Pressing the Import Options button will open the Spectral Import Options dialog box (Figure 26). This is where information about the m/z values of peaks is controlled.
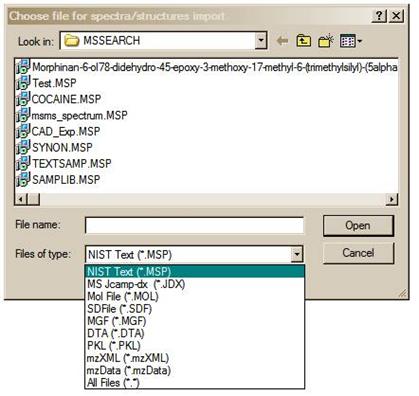
Figure 24. File Selection dialog box with some of the available file types. MSP and JDX files are spectra, and MOL files are structures.
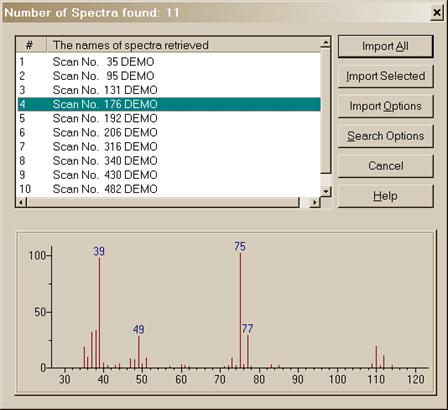
Figure 25. Spectrum Import dialog box with a single spectrum from a multiple spectrum file highlighted and displayed in a bar‑graph format.
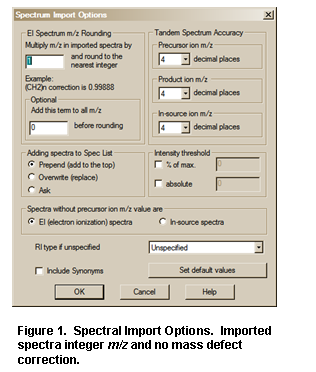 The two fields on the upper left of
the Spectral Import Options dialog box are used to make mass defect
corrections. The difference in the integer nominal mass and the exact mass of
an isotope is the mass defect. The most significant effect of mass
defect is seen on the integer mass of hydrocarbons due to the mass defect
of hydrogen. The integer mass of the 1H isotope is 1. The exact
mass of this isotope is 1.00725. The C50H100 ion has a
nominal mass (the sum of integer mass of the most abundant naturally occurring
isotope of each element times the number of atoms of those elements) of 700,
(50 ´ 12) + (100 ´ 1). The integer value for C50H100 based on a
calculation of the exact mass of each of these elements is 701 (12.000000 ´ 50) + (100 ´ 1.007825) =
700.7825. The mass defect for C50H100 is 0.7825,
which is equal to about 100 millimass units per 100 mass units. The MS
Search Program deals with only integer mass values when searching the NIST
14 MS Library. Spectra are imported with integer m/z values;
therefore, spectra recorded with observed values of m/z, which are not
integers and have not been corrected for mass defect, must have a mass
defect correction applied in order to be properly searched against the NIST
14 MS Library or another library that contains only integer m/z
values.
The two fields on the upper left of
the Spectral Import Options dialog box are used to make mass defect
corrections. The difference in the integer nominal mass and the exact mass of
an isotope is the mass defect. The most significant effect of mass
defect is seen on the integer mass of hydrocarbons due to the mass defect
of hydrogen. The integer mass of the 1H isotope is 1. The exact
mass of this isotope is 1.00725. The C50H100 ion has a
nominal mass (the sum of integer mass of the most abundant naturally occurring
isotope of each element times the number of atoms of those elements) of 700,
(50 ´ 12) + (100 ´ 1). The integer value for C50H100 based on a
calculation of the exact mass of each of these elements is 701 (12.000000 ´ 50) + (100 ´ 1.007825) =
700.7825. The mass defect for C50H100 is 0.7825,
which is equal to about 100 millimass units per 100 mass units. The MS
Search Program deals with only integer mass values when searching the NIST
14 MS Library. Spectra are imported with integer m/z values;
therefore, spectra recorded with observed values of m/z, which are not
integers and have not been corrected for mass defect, must have a mass
defect correction applied in order to be properly searched against the NIST
14 MS Library or another library that contains only integer m/z
values.
The mass defect factor can be entered in the box “Multiply m/z in imported spectra by”. The factor for hydrocarbons based on a general formula of –(CH2)n– is 0.99888. When dealing with spectra that have significant mass contribution from multiple atoms of chlorine or bromine (both of which have a significant negative mass defect), a different factor will have to be calculated. In this case, it may be desirous to use the field preceded by “Add following term [value] to all m/z values before rounding”.
The “Adding spectra to Spec List” box allows the permanent specification of the action when importing spectra. Choosing Prepend will always add to the current Spec List, Overwrite will always replace the current Spec List. Ask was the only option prior to version 2.2 of the search program.
Intensity threshold allows you to remove low intensity peaks from the spectrum as it is being imported. This can be done in terms of relative peaks – for example 0.1% would keep peaks to 1 part in 1000 of the base peak. Absolute will simply remove data below a user specified threshold.
Spectra without precursor ion m/z values are assumed to be EI data by default, but can be changed to be In-source/EI with accurate ion m/z spectra.
RI data in input spectra are not normally characterized by type of column. If you want to specify a particular column type for your data you may do so here.
Finally Synonyms may be included or excluded
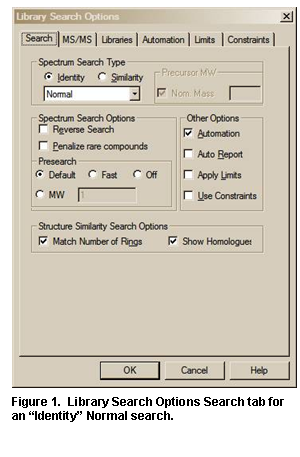 Library Search Options
Library Search Options
Selecting Library Search Options on the Options menu from any tab or selecting the Library Search Options button on the button bar of the Lib. Search tab will display a multiple-tab Library Search Options dialog box (Figure 27).
A number of different types of spectral comparison routines for choosing the best matching spectra are available. The routine to be used is selected from the Library Search Options dialog box’s Search tab.
The two search types are Identity (with three options) and Similarity (with four options). These different searches and their options are explained in the Help Screens (topic Spectrum Search Type).
The Search tab of the Library Search Options dialog box also has a Structure Similarity Search Options selection. This has the Match Number of Rings check box. When selected, the search of compounds that have rings will be constrained to only spectra produced by compounds with a similar structure and the same number of rings as the spectrum being searched. The Show Homologues selection is self-explanatory.
MS/MS
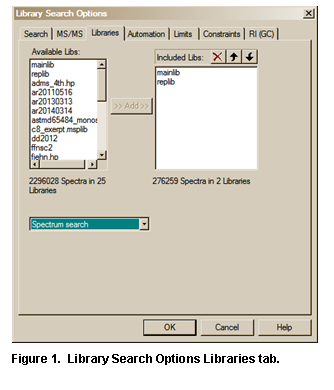 The MS/MS tab permits
selection of the m/z tolerances for the precursor and product ions in
the MS/MS and
In-source HiRes Search, the option to ignore the precursor ion, and access to a set of options specific to
peptides.
The MS/MS tab permits
selection of the m/z tolerances for the precursor and product ions in
the MS/MS and
In-source HiRes Search, the option to ignore the precursor ion, and access to a set of options specific to
peptides.
Libraries
The Libraries tab (Figure 28) of this dialog box is used to select the libraries to be included in the Library spectrum Search. As many 127 libraries can be searched at the same time. This includes the NIST 14 Main Library (mainlib) and the NIST 14 Replicates Library (replib) and multiple user libraries as well as other commercially available libraries from sources other than NIST that are in the NIST MS Search library format. By default, three other NIST 14 Libraries are also installed: the nist_ri, nist_msms, and nist_msms2. This means that there is space to accommodate an additional 122 user libraries. If more MS libraries need to be accommodated, the less used ones can be moved to another folder. To add libraries to be searched in the Available Libs pane of the dialog box, highlight them and click on the Add button. To exclude libraries, highlight them in the Included Libs pane and then click on the X button above the upper right portion of this pane. The order in which libraries are searched can be changed by highlighting the library to be moved in the Included Libs pane and clicking on the Up or Down arrows in the upper right portion of this pane. Each click will move the library one position. The order in which libraries are searched is not important because the Hit List has the 100 Highest Match Factor Hits regardless of what library they came from.
Below the Available Libs and Included Libs panes is a drop-down list box. This box allows for switching between the lists of libraries to be included in Structure searches or Spectra searches. These two different types of searches can have different sets of libraries.
Retention Index Data
The retention index search options are generally only available when the search spectrum has a retention index (RI) value in the submitted spectrum. The Ri value is taken either from a line in the MSP file of the format
RI=value
or from the corresponding text in comment section of the MSP file.
The use of the RI data is controlled by the RI(GC) tab in the Library Search Options dialog box (Figure 29). If the “Use Retention Indices…” box is checked, then the program will compare the RI data in the submitted spectrum to the RI value in the library spectrum. The comparison will only be made if the type of column for the search spectrum and the library spectrum are the same.
 In general the data will not have the column type specified
so it the specification can be made at the time the data is imported.
Alternatively, if the search spectrum column type is not explicitly stated, the
program will assume that it is the type specified in the box “Select RI
column Type for Searching or Displaying”.
In general the data will not have the column type specified
so it the specification can be made at the time the data is imported.
Alternatively, if the search spectrum column type is not explicitly stated, the
program will assume that it is the type specified in the box “Select RI
column Type for Searching or Displaying”.
The normal mode of operation will restrict the search to data of the same column type. However, since the two non-polar types are somewhat similar, the program has the option to allow the use of the other polar type if the specified type is not present. NOTE: If there is both standard (DB1) and semi-standard(DB5) non polar data, and you have specified the DB5 data, it will be used. The DB1 data will only be used if a specific library spectrum does not have data for DB5. No such option is available for the polar column data. To select the option to use the alternate non-polar type check the box next to “Use other non-polar column…” below the Select RI Column box.
When indexing a user library the option exists to specify the column type. If it is not specified and the column type of the individual RI values for spectra in the library are not specified, then the column type will be set to “User” or unspecified. In this case you must check the last option to ensure that the RI data in the user library is correctly processed.
A brief note on the values of RI tolerance and RI penalty rate: The RI variation within a set of data taken using basically the same protocols and generally similar matrices will vary by a very small amount, typical standard deviations are in the range of 2-3 RI units. The median variation (that is the median of the uncertainty (median absolute deviation) of the data in the NIST RI library is over 6 RI units. Thus the data in the NIST library may be at greater variance from your experimental data. For this reason the recommended RI tolerance is fairly high and the penalty rate is low.
The use of both non-polar data sets, while expanding the range of RI data must be done with care. Looking at all the data in the collection where there are both DB1 and DB5 data, the DB5 data is about 0.74% greater than the DB1, that is RI(DB5) =1.0074*RI(DB1). This is a small variance, but at a RI of 2000, it corresponds to a difference of 14.8. In addition the scatter in the correlation data is fairly large. If the RI tolerance is 10 and the wrong column type is being used since the correct one is not available, there may be too much penalty.
Automations
When a Library spectrum Search or a Structure Search is carried out on a spectrum or a structure that is imported from a file or third-party program such as an instrument data system or drawing program, a search can be automatically launched. Automatic searching and/or printing can be turned on or off from the Search tab of the Library Search Options dialog box by the respective selection or deselecting of the appropriate check box (Figure 27 and Figure 30).
 The Automation tab of the Library
Search Options dialog box is used in conjunction with these automated
searches (Figure 30). Check box Automatic
Search On (Figure 30) has exactly same functionality as Automation
check box on Figure
27: turns on automatic
searching. Selecting or deselecting this check box on either tabs
toggles the check box on the other tab.
The Automation tab of the Library
Search Options dialog box is used in conjunction with these automated
searches (Figure 30). Check box Automatic
Search On (Figure 30) has exactly same functionality as Automation
check box on Figure
27: turns on automatic
searching. Selecting or deselecting this check box on either tabs
toggles the check box on the other tab.
Search results can automatically be printed whether or not the search is automatically launched. Check box Auto Report (Figure 27) turns on this feature. “Number of Hits to Print” field is located in the Automation tab (Figure 30). In addition, the report in the same format can be printed for any displayed search result by selecting Print Auto Report from the File menu.
The top part of this dialog box pertains to the automated printing of a search result. The format for the Auto Report is a sample spectrum at the top of a page followed by the spectra of a specified number of Hits in the “Number of Hits to Print” numeric entry box (2 by default). If the Include Spectrum Plot in Report check box is selected, there will be three spectra per page. Page one will have the sample spectrum at the top followed by the first two Hits. Selecting the Draw Structures in Plots check box will result in the structure associated with a spectrum being printed in the plotted spectrum.
If the Apply Maximum Spectrum Length check box is selected, a number can be entered for the maximum m/z value to appear on the spectrum’s abscissa. If peaks are present at higher m/z values, the abscissa will have a right-pointing arrowhead at its right end. When selected, the Return Focus to Caller Upon Completion check box will cause a program that brought the NIST MS Search Program into focus if already running, or started if not already running, to again be visible after the search.
Limits
The function of the limits of the starting and ending m/z values of a Library spectrum Search and the lowest value of a mass spectral peak that will be considered in a search can be set. An explanation of the limits that can be used to constrain a search may be found in the Help file by selecting the Help button on the Limits tab of the Library Search Options dialog box.
Constraints
A Library spectrum Search can be constrained by a number of factors such as what elements (and how many atoms of each) must be present in the compounds that produce retrieved spectra, the molecular weight (nominal mass) range of Hits, whether or not the compounds that produce retrieved spectra are in various other libraries, what m/z value peaks must be present (along with a range of their relative intensity) in retrieved spectra, what name fragments must be associated with retrieved spectra, what text strings are in the “Comments/Contributor” field of a retrieved spectra, and new to v.2.0g of the MS Search Program: an Exact Mass Constraint used instead of a molecular weight range. These constraints are explained in the Help file, which is displayed by selecting the Help button on the Constraints tab of the Library Search Options dialog box.
The Search Menu (displayed by Selecting Search on the Menu Bar)
The Other Search tab’s options are:
|
Option |
Description |
Tab View |
|
MW (Nominal Mass) |
Finds spectra in selected libraries of compounds having specified (nominal mass) molecular weight (excludes replib). |
Other Search |
|
Exact mass |
Finds spectra in selected libraries of compounds having an exact mass corresponding to specified accurate mass with a specified variance (excludes replib). |
Other Search |
|
Formula (Elemental Composition) |
Finds spectra in selected libraries of compounds with specified formula (excludes replib). |
Other Search |
|
Any Peaks |
Finds spectra in selected libraries based on user-specified peaks. |
Other Search |
|
Sequential Method |
Searches selected libraries using a set of constraints provided by the user. |
Other Search |
|
CAS Number |
Finds spectra of the compounds that have the entered CAS registry number. |
Other Search |
|
ID Number |
Finds spectrum (or spectra) in selected library having entered ID number (or range of ID numbers). |
Other Search |
|
NIST Number |
Retrieves a spectrum with unique NIST Mass Spectral Data Center’s Archive ID number (in mainlib and replib only). |
Other Search |
|
Name |
Allows entering a character string to display spectra beginning with name of a compound. |
Names |
These options (with the exception of the Name search) are also displayed and may be selected from the drop‑down list box on the further most left side of the Other Search tab (to the right from the binocular button). Selecting the binocular button starts the currently displayed search in the drop-down list box.
Each Search has its own dialog box. When the MW (Nominal Mass) Search, Formula Search, or Sequential Method Search is selected, the dialog box opens to the Constraints tab (Figure 31). The MW (Nominal Mass) Search and the Formula Search dialog boxes have specific entry fields for entering a nominal mass and elemental composition, respectively. Certain obvious constraints are not available (appear grayed in the constraints list and cannot be selected), such as “MW range” and “Exact Mass” in the MW (Nominal Mass) Search dialog box and “Element Values” and “Elements Present” in the Formula Search dialog box. These three dialog boxes have two tabs. The other tab is the Options tab (Figure 32), which allows for the selection of the libraries to be searched. Even though it is present, the replib cannot be selected for a MW (Nominal Mass), an Exact Mass, or a Formula Search. Spectra from the replib for these searches automatically appear as replicate spectra in the Hit List.
The maximum number of Hits allowed in the Hit List can be specified in the Options tab of the Sequential Method Search. There is also a limit on the maximum number of Hits that can be in a Hit List of an Any Peaks Search (settable in the Peaks tab). The Lib. Search Hit List is limited to 100 Hits. There is no limit to the number of Hits that can be in a MW (Nominal Mass), an Exact Mass, or a Formula Search.
Search by Any Peaks
Figure 31. Formula
Search dialog box with Use Constraints selected and a Tag in Constraints
entered to yield only spectra contributed by Carl Djerassi.
Select Any peaks
from the Other Search tab’s Search menu or the drop-down list
box. This will display the Any Peaks Search dialog box (Figure 33). Click the “m/z” field
(located below the m/z drop-down list box). The blinking vertical text
cursor will now appear in “m/z” field. “Normal” will be displayed in
“Type” field. Type 82 and press <Enter>. This moves the cursor
to the “From” field. Type 70 and press <Enter>. This
moves the cursor to the “To” field. Type 100 and press <Enter>.
This will enter the peak with its type and intensity range. The cursor is now
in the “m/z” field.
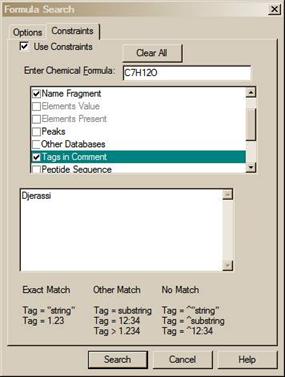
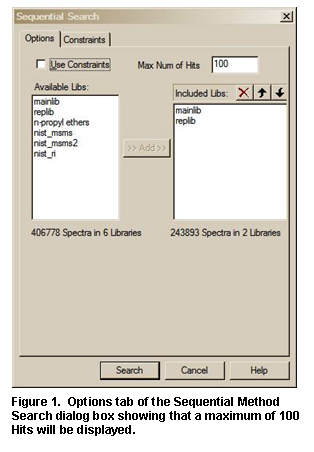 Note that the option of the total number of hits will
default to 100, but can be set to a maximum of 1000.
Note that the option of the total number of hits will
default to 100, but can be set to a maximum of 1000.
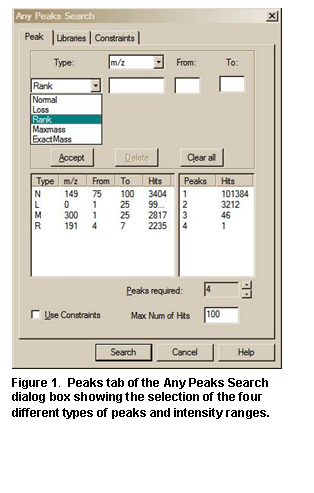 Put the mouse pointer on the ê on the right side of the “Type”
field and click the left Mouse button. Put the Mouse pointer on “Loss” in the
displayed menu and click the left Mouse button again. This will cause the
cursor to reappear in the “m/z” field. “Loss” will be displayed in the
“Type” field. The first peak entered is an observed peak in the mass spectrum,
and it is possible that it is the base peak. The second peak entered
represents an ion formed by a loss from the molecular ion. Type 31 and
press <Enter>. Type 5 and press <Enter>. Type 25 and press
<Enter>. The retrieved spectra must have a peak at 31 m/z
units lower than the molecular peak that exhibits intensity between 5% and 25%
of full scale. This reduces the number of spectra in the NIST/EPA/NIH Mass
Spectral Main and Replicates Libraries that match the entered
criteria from 1796 for the first peak to 122 that have both peaks
Put the mouse pointer on the ê on the right side of the “Type”
field and click the left Mouse button. Put the Mouse pointer on “Loss” in the
displayed menu and click the left Mouse button again. This will cause the
cursor to reappear in the “m/z” field. “Loss” will be displayed in the
“Type” field. The first peak entered is an observed peak in the mass spectrum,
and it is possible that it is the base peak. The second peak entered
represents an ion formed by a loss from the molecular ion. Type 31 and
press <Enter>. Type 5 and press <Enter>. Type 25 and press
<Enter>. The retrieved spectra must have a peak at 31 m/z
units lower than the molecular peak that exhibits intensity between 5% and 25%
of full scale. This reduces the number of spectra in the NIST/EPA/NIH Mass
Spectral Main and Replicates Libraries that match the entered
criteria from 1796 for the first peak to 122 that have both peaks
The next peak will be a “Maxmass” peak. Click on the ê on the right side of the “Type” field. Click on “Maxmass”. Type 303 and press <Enter>. Type 1 and press <Enter>. Type 50 and press <Enter>. There will only be eight spectra that contain all three peaks of the designated type and over the designated intensity range.
NOTE – USER LIBRARIES ARE NOT SEARCHED USING ANY PEAKS EXACT MASS
The Any Peaks Search can also perform searches on Exact Mass of the fragments. However, this search mode only works on the NIST libraries; it will not function on user libraries even if the user library has been indexed by exact mass. The exact mass indexing of the user libraries done through the Tools/(Re)Index Exact Mass only indexes the precursor or molecular ion based on the formula.
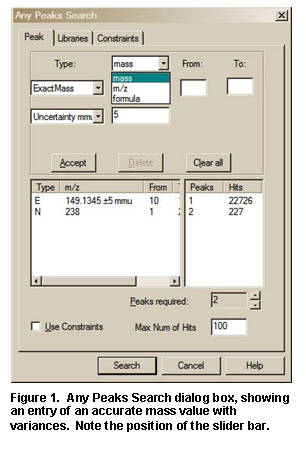 Figure 34 is the Any Peaks Search dialog box set to accept an accurate mass
value with variance. It is important to remember that the “Exact Mass” Any
Peaks Search is only valid for monoisotopic peaks (all atoms of all
elements in the ion represented by the peak are the most abundant isotopes).
Do not try to enter values for peaks that contain higher mass isotope of
chlorine (37Cl), bromine (80Br), or other elements. An
accurate mass (Mass), an m/z value (adjusted for electron mass),
or an elemental composition (Formula) can be entered.
Figure 34 is the Any Peaks Search dialog box set to accept an accurate mass
value with variance. It is important to remember that the “Exact Mass” Any
Peaks Search is only valid for monoisotopic peaks (all atoms of all
elements in the ion represented by the peak are the most abundant isotopes).
Do not try to enter values for peaks that contain higher mass isotope of
chlorine (37Cl), bromine (80Br), or other elements. An
accurate mass (Mass), an m/z value (adjusted for electron mass),
or an elemental composition (Formula) can be entered.
The uncertainty can be in millimass units (mmu) or parts-per-million (ppm). If this field is left blank, an uncertainty of 0.5 less than the last digit entered for the mass is assumed. In this case the uncertainty always is a 0.5 value; i.e., the uncertainty for 120.00 is ±0.005 and 120.000 is ±0.0005.
To enter a chemical formula, select formula in the drop-down list box above the first entry field (this is possible only if “Exact Mass” type has been selected.) If the “Uncertainty” field is left blank, the accuracy of searched mass is exactly the same as the accuracy of the mass saved in the library. It is very rare the exact mass of two different elemental compositions is the same; therefore, there is a high probability only the spectra where the peak was interpreted with the same elemental composition will be found. Molecular ion peaks in the mass spectrum will give very good results when used for the Any Peaks Search Exact Mass option; however, caution must be taken when using the accurate mass of fragment ions. The exact mass values in the NIST DB are calculated from the elemental compositions of ions predicted by MS Interpreter where peaks are present in the mass spectrum. More than one exact mass may be saved in a library for a peak in a mass spectrum of a compound. For instance, both CO and C2H4 may be assigned to the same peak with m/z 28. Searching for these two ”Formula” would produce a number of Hits containing both peaks. These Hits correspond to mass spectra where the peak at m/z 28 was ambiguously interpreted. The Any Peaks criteria can be used in the Sequential Method Search or as a constraint with a MW (Nominal Mass), Exact Mass, Formula Search or with a Spectrum Search; however, the “Exact Mass” Peak type is not available for these searches. When Peaks is used as a Constraint, relative intensity can be specified. In this case, the subsequent peaks have intensities relative to the first peak. Thus, if the first peak has an intensity between 50 and 100% of base, the second peak can be specified to have an intensity of between 40 and 200% of the first peak. This is especially important when using SIM (selected ion monitoring) searches and wanting to understand the rates of false-positives. Radio buttons appear at the bottom of the Peaks Constraints dialog box to switch back and forth between “Absolute” and “Relative” (1st Peak is Special !).
Exact Mass Search
Exact Mass Search can be used directly to search the NIST libraries. In addition, Exact Mass (alternately with nominal mass) can be used as a Constraint in User Spectra Searches (Lib. Search tab), Any Peaks Search, and Sequential Search (Other Search tab).
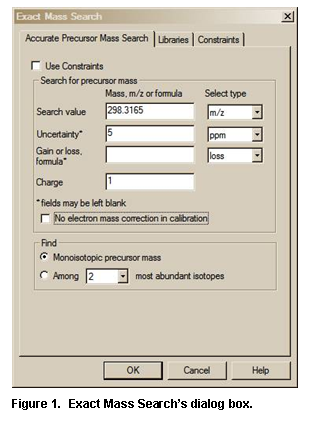 NOTE – THE EXACT MASS CONSTRAINT IS FOR MOLECULAL OR
PRECURSOR ION MASS
NOTE – THE EXACT MASS CONSTRAINT IS FOR MOLECULAL OR
PRECURSOR ION MASS
 Select Exact Mass from the
drop-down list box on the upper left of the Other Search tab. The
three-tab dialog box shown in Figure 35 will be
displayed. The exact mass value to be searched is entered into the “Search
value” field on the Accurate Precursor Mass Search tab of the dialog
box. The “Type” of the value is selected from the drop-down list box next to
the “Search value” field. All of the fields in the dialog box are explained in
detail by selecting the dialog box’s Help button. The Libraries
tab allows the selection of the libraries to be searched. Like the MW (Nominal
Mass) Search and the Formula Search, the NIST 14 replib
is not available for searching. The Constraints tab allows the Exact
Mass Search to be constrained by presence in other databases, name
fragment, Tags in the “Comments/Contributor” field, elements present, and
element values. In addition, the formula or the formula for the neutral loss
can be entered. In this case the program will calculate the exact mass and use
the uncertainty (if provided).
Select Exact Mass from the
drop-down list box on the upper left of the Other Search tab. The
three-tab dialog box shown in Figure 35 will be
displayed. The exact mass value to be searched is entered into the “Search
value” field on the Accurate Precursor Mass Search tab of the dialog
box. The “Type” of the value is selected from the drop-down list box next to
the “Search value” field. All of the fields in the dialog box are explained in
detail by selecting the dialog box’s Help button. The Libraries
tab allows the selection of the libraries to be searched. Like the MW (Nominal
Mass) Search and the Formula Search, the NIST 14 replib
is not available for searching. The Constraints tab allows the Exact
Mass Search to be constrained by presence in other databases, name
fragment, Tags in the “Comments/Contributor” field, elements present, and
element values. In addition, the formula or the formula for the neutral loss
can be entered. In this case the program will calculate the exact mass and use
the uncertainty (if provided).
The Exact Mass Search is not restricted to the NIST/EPA/NIH Libraries; however, user or other non-NIST libraries must first be indexed according to exact mass. Select Tools from the Main Menu and then select (Re)Index Exact Mass. This will result in a dialog box being displayed (Figure 36). The time to index a library will depend on the size of the library. All NIST 14 Libraries are already indexed according to exact mass.
After the desired libraries have been indexed (this has to be done only once), an Exact Mass Search or another Search using Exact Mass as a Constraint can be carried out.
Search by Name - Browse
The data in a particular library can be searched or browsed using the incremental name search (the Names tab). As a name is entered, the program finds the next name that fits all of the text typed to that point. The names search works over not only the primary name but all synonyms. This means that common names for chemicals can be quickly searched. Some common compounds have a very large number of synonyms. Once the name is found highlight the desired name, and the display in the Plot/Text and Plot Windows will be refreshed with the appropriate display for the compound.
Use the Mouse pointer to place the blinking vertical text cursor in the “Name” field at the top of the Name Search tab. To search for the mass spectrum of a plant growth regulator, GIBBERELLIN A9, begin typing the name. Stop after entering each letter. After the letter N has been typed, the 13th name in the Hit List pane is the target. Names may also be pasted from the Windows Clipboard into the “Name” field by simultaneously pressing keys Shift and Insert or using the RMB menu’s Paste selection.
NOTE: This feature can be used in a user library to give synonyms such as “LabBook05-Pg6” or more simply “B05-P06”, this allows the data to be quickly found by reference to the original experiment.
Sequential Method
This method allows every spectrum in the selected libraries to be subjected to a created search criteria. The criteria that are set in the Constraints section govern the Hit List returned. In this search method it is possible to use not only absolute but also only relative intensities of peaks. This can be an important tool for examining the risk of false positives when using SIM searches.
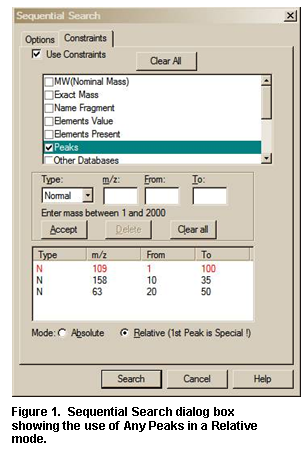 In the example shown in Figure 37, the peak at 109 is allowed to be
present in a range of 1 to 100% of the base peak. The subsequent peaks (158
and 63) are sought as a fraction of the 109 peak, regardless of the intensity
of the 109. If the peak with m/z 109 is 15% of the intensity of the
base peak and the peak with m/z 158 is 5% of the intensity of the base
peak (i.e., 33% of the 109) it would be accepted. This kind of analysis can
give important insights into to risk of false positive matches when using SIM
(selected ion monitoring) in GC/MS experiments.
In the example shown in Figure 37, the peak at 109 is allowed to be
present in a range of 1 to 100% of the base peak. The subsequent peaks (158
and 63) are sought as a fraction of the 109 peak, regardless of the intensity
of the 109. If the peak with m/z 109 is 15% of the intensity of the
base peak and the peak with m/z 158 is 5% of the intensity of the base
peak (i.e., 33% of the 109) it would be accepted. This kind of analysis can
give important insights into to risk of false positive matches when using SIM
(selected ion monitoring) in GC/MS experiments.
In SIM the entire spectrum is not seen so the relative intensities of the ions observed are the only data that can be extracted from the data file.
NOTE: All of the other methods of searching are also present in the Sequential Method. The elements search can also be interesting if the number of compounds in all of the libraries that have at least one mercury atom in the molecule needs to be determined. On a modern computer with a large amount of memory these searches do not need to take very long. Typically any sequential search will be faster for subsequent similar searches because much of the data file will be cached in the computer memory.
Basis for Interpretation of the Library Search Results
When an Identity Search with an unknown spectrum (Library spectrum Search) is performed, the results will be associated with three numbers for each spectrum shown and one value for the search. These three numbers are 1) a Match Factor for the unknown and the library spectrum (direct match), 2) a match factor for the unknown and the library spectrum ignoring any peaks in the unknown that are not in the library spectrum (Reverse Match), and 3) a Probability value (column headings in the Lib. Search Hit List: Match, R. Match and Prob., respectively). The first two numbers are fairly straightforward. Each is derived from a modified cosine of the angle between the spectra (normalized dot product). A perfect match results in a value of 999; spectra with no peaks in common result in a value of 0. As a general guide, 900 or greater is an excellent match; 800–900, a good match; 700–800, a fair match. Less than 600 is a very poor match. However, unknown spectra with many peaks will tend to yield lower Match Factors than similar spectra with fewer peaks. The Probability value and the value for the search (InLib, located in the lower right corner of the Hit List pane and on the Title Bar of the Lib. Search tab) require more explanation.
The Probability value for a Hit is derived assuming that the compound is represented by a spectrum in the libraries searched. It employs only the differences between adjacent Hits in the Hit List to get the relative probability that any Hit in the Hit List is correct. This value is derived from an analysis of the results of searching the NIST/EPA/NIH Main Library with a set of replicate spectra (given in the Replicates Library). The relative probability of each of the Hits requires only the difference values because the total probability of the compound being in the searched libraries is assumed to be one. The other factor (InLib) is a measure of the probability of the compound actually being in the searched libraries. This value is also derived from the same set of replicate spectral searches. In this case, the correct compound was ignored in the Hit List; and the difference between the Hit List, with and without the correct compound, is parameterized. The parameters are the maximum value of the match and the largest single difference among the top 20 Hits. If the first Hit has a high Match Factor (>900) and the next Hit has a Match Factor of 800 or less, the probability of the compound being correctly identified is very large and the probability of the compound being in the searched library is large (the number of “Hits vs. their Match Factors” is displayed in the Hit Histogram pane located just above the Hit List, see Figure 3).
Like all statistical results, these probability calculations rely on the data sampled. For example, if a compound that has very few similar mass spectra will be identified in a more definitive way (a higher Probability and InLib value). Using examples from the Replicates Library and searching the Main Library using a compound like 'folpet', will result in a high probability for the first Hit and a high InLib value. In contrast, if replicates of 'cyclohexanol' are searched, the results will be much lower Probabilities and InLib value; and, in some cases, the best Match is not even the correct compound. This reflects the fact that there are very few compounds that have mass spectra similar to 'folpet'; but there are a number of compounds that have very similar mass spectra to 'cyclohexanol', and the ability of any search system to distinguish between these similar spectra is limited. In many cases, the best that the search can do is to provide a class of compounds that have similar mass spectra and, usually, similar structures.
The values of the InLib parameter are meant as guideposts. Generally, any positive value is acceptable. Values greater than approximately 300 usually mean that the spectrum is nearly unique. Negative values below 200 are generally a warning that the spectrum is not identified. Note that negative values will occur when there are a large number of compounds with similar spectra. In these cases, the difference between the Match Factors for different spectra is very small, and the search cannot be assured of providing the correct unique answer. Usually in these cases, especially when Match Factors are high, it will provide very good guidance on the structure of the molecule.
For a complete discussion of the methods used in assessing the probabilities, see Stein, S.E. “Estimating Probabilities of Correct Identification from Results of Mass Spectral Library Searches” J. Am. Soc. Mass Spectrom. 1994, 5, 316–323.
Substructure Information
The Substructure Information utility is used in the evaluation of a Hit List using the NIST/EPA/NIH Mass Spectral Library because each spectrum is indexed according to the substructures that make up the structure of the compound that resulted in the spectrum. Substructures are functional groups like carbonyls, methoxys, methyl ester moieties, etc.; the number of rings and double bonds; and heteroatoms.
The Substructure Information is most useful when the Hit List is the result of one of the four Similarity Searches (Simple, Hybrid, Neutral Loss, MS/MS in EI). This utility is called from the Main Menu Bar’s Tools menu.
Similarity Searches are carried out when it is believed that a spectrum of the unknown compound is not in any of the searched libraries. Spectra similar to the searched spectrum make up the Hit List. There is no Prob column in the Hit List and an InLib value is not determined. Not only is the utility useful in determining the structure of an unknown based on its EI mass spectrum, it has proved to be very informative in the analysis of collision-induced dissociation (CID) spectra obtained from ions produced by soft ionization LC/MS techniques such as APCI, APPI, and electrospray. This use is described in more detail below.
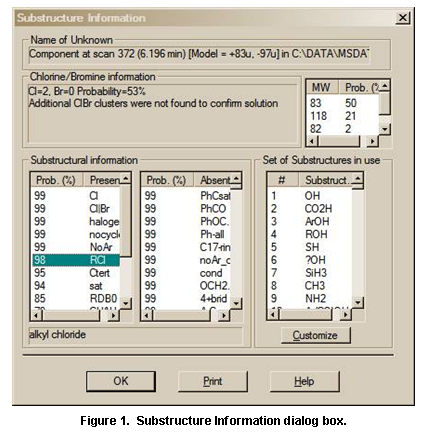 The Substructure Information
dialog box (Figure 38) has the name of the spectrum that
was used to create the Hit List displayed at the top. Just below the
“Name of Unknown” field in this dialog box is the “Chlorine/Bromine
information” field, which gives the probabilities and numbers of atoms of
chlorine and/or bromine present (useful regardless of the method by which the
spectrum was obtained). The result of molecular weight estimation from the Hit
List is presented on the right side of the dialog box (useful for EI
spectra, especially when no molecular ion peak is observed). There are two
panes below this in an area labeled Substructure information: Prob.
Present and Prob. Absent. These two panes contain a list of
abbreviations for the substructures preceded by a number, which is the percent
probability for the presence or absence of the substructure. If one of the
abbreviations is not understood, highlight it by using the Mouse and a detailed
explanation appears at the bottom of the Substructural information area.
The Substructure Information
dialog box (Figure 38) has the name of the spectrum that
was used to create the Hit List displayed at the top. Just below the
“Name of Unknown” field in this dialog box is the “Chlorine/Bromine
information” field, which gives the probabilities and numbers of atoms of
chlorine and/or bromine present (useful regardless of the method by which the
spectrum was obtained). The result of molecular weight estimation from the Hit
List is presented on the right side of the dialog box (useful for EI
spectra, especially when no molecular ion peak is observed). There are two
panes below this in an area labeled Substructure information: Prob.
Present and Prob. Absent. These two panes contain a list of
abbreviations for the substructures preceded by a number, which is the percent
probability for the presence or absence of the substructure. If one of the
abbreviations is not understood, highlight it by using the Mouse and a detailed
explanation appears at the bottom of the Substructural information area.
Just below the molecular weight information is the Set of Substructures in use pane with a list box and a Customize button. The list box displays all the substructures in the current set.
The Customize button, when
selected, displays the Customized set of Structures dialog box (Figure 39). This dialog box is used to create
a subset of the substructures to be identified. The list of all possible
substructures is displayed in the Full set of structures pane. The list
of substructures  included in the present file is shown
in the Customized set pane. The first time a customized list is
created, both panes will contain the same list. Substructures may be omitted
from the Customized set pane or added from the Full set of structures
pane. Either action occurs on the structures highlighted by Mouse action in
the panes by selecting the appropriate button ß Omit ß or à Add à. The usual Windows conventions for
selecting multiple items apply. After selecting the first substructure of the
list, hold down the <Shift> key while selecting the last substructure.
This will select all substructures between the first and last. Multiple
noncontiguous substructures can be selected by holding down the <Ctrl>
key while selecting each desired substructure.
included in the present file is shown
in the Customized set pane. The first time a customized list is
created, both panes will contain the same list. Substructures may be omitted
from the Customized set pane or added from the Full set of structures
pane. Either action occurs on the structures highlighted by Mouse action in
the panes by selecting the appropriate button ß Omit ß or à Add à. The usual Windows conventions for
selecting multiple items apply. After selecting the first substructure of the
list, hold down the <Shift> key while selecting the last substructure.
This will select all substructures between the first and last. Multiple
noncontiguous substructures can be selected by holding down the <Ctrl>
key while selecting each desired substructure.
The Save and Open buttons allow the saving and retrieving of customized substructure sets. A temporary set is created by making modifications to the Customized set pane and selecting the OK button. This dialog box, as well as the Substructure Information dialog box, has a Help button, which displays a context‑sensitive Help screen.
The algorithms used in the substructure identification are described in the Help screen. They are based on developments at NIST (Stein, S.E. “Chemical Substructure Identification by Mass Spectral Library Searching” J. Am. Soc. Mass Spectrom. 1995, 6, 644–655).
Example of the Use of Substructure Information in Making Determination about an Unknown Mass Spectrum obtained by using CID
The mass spectrum in Figure 40 of a compound with the structure shown inserted in the spectrum was obtained using collision-induced dissociation (CID) on a protonated molecule with m/z 248. An EI in MS/MS Similarity Search was performed against the NIST 14 mainlib with m/z 248 designated as being the precursor ion. This type of search is provided to allow for the use of the vast amount of information in the EI Library with spectra that are produced by LC/MS CID. The EI in MS/MS Similarly Search has been optimized for this purpose. The resulting Hit List was evaluated using the Substructure Identification utility. Based on this evaluation, it was determined that there was a 90% probability that the compound contained one or more atoms of oxygen; a 90% probability that there was a carbonyl group; an 89% probability that there was a heterocyclic ring; and an 88% probability that there was an aromatic ring; but it also revealed a 99% probability that there was not a phenyl carbonyl or a methoxy phenyl group, and a 90% probability that there was not a heterocyclic ring that contained oxygen. Remember, it is just as important to know what is absent as it is to know what is present. The evaluation revealed that there was an 89% probability that there were one or more atoms of nitrogen and an 86% probability that there was a heterocyclic ring with an atom of nitrogen. Continued evaluation of the Prob. Present and Prob. Absent panes in the Substructure Identification dialog box revealed other possible substructures to be present and eliminated others by their high-absence probabilities.
This particular use of the MS/MS in EI Similarity Search in conjunction with the Substructure Identification has not been extensively tested by NIST; however, anecdotal evidence has shown this to be an effective combination and should be considered when trying to determine the structure of an unknown using CID data. In the example cited above, if the unknown was believed to be a metabolite of some known substances, the information gained from this technique could well result in identification.
Use with Instrument Data Systems
The features described in the AUTOMATIONS section of this manual were implemented aid the use of the NIST MS Search Program when used with proprietary mass spectral data systems.
Location of the NIST MS Search Program
It may be necessary to determine the location of the NIST MS Search Program files and the location of the NIST MS Search working directory. These may be found in the [NISTMS] section of the WIN.INI file located in the %windir% directory. Below is a typical example of this section:
[NISTMS]
Path32=C:\NIST14\MSSEARCH\
WorkDir32=C:\NIST14\MSSEARCH\
Amdis32Path=C:\NIST14\AMDIS32\
AmdisMSPath=C:\NIST14\AMDIS32\
The string beyond “Path32=” is the path to MS Search Program’s folder (location of the executable files); the string beyond “WorkDir32=” is the path to the MS Search Program’s working directory (location of the history, Hits, etc.). Two other items refer to AMDIS and its connection to the MS Search.
Implementation
If the mass spectrometry data system being used is capable of exporting spectra to the NIST format (described in the USER DATABASES section of this manual) and a Windows program can be executed from within the data system, this feature is available. Implementation of this feature has been provided by some manufacturers on their data systems.
The command string used to start or bring into focus the NIST MS Search Program for Windows is:
C:\NIST14\MSSEARCH\NISTMS$.EXE<space>/instrument
assuming the program is in the C:\NIST14\MSSEARCH directory; if it is in some other directory, make the appropriate substitution.
Two locator files are required to use the automation features:
1. The first locator file must be in the MS Search Program’s working directory, have the name AUTOIMP.MSD, and contain the text string describing the name and location of the second locator file. For example:
C:\MYMSDS\FILESPEC.FIL
where: C:\MYMSDS is the folder where the file, FILSPEC.FIL, resides.
This file can be created with any text editor.
2. The second locator file is created each time spectra are saved to a separate file to be imported by the MS Search Program. This second locator file contains the text string that describes the name and location of the text file containing the spectrum (or spectra) in the NIST Text file format described later. It is possible to state whether the spectra currently in the NIST MS Search Program’s Spec List pane are to be added to or replaced by the automatic import. An example of the contents of the second locator file is:
C:\MYMSDS\DATA\DATAFILE.TXT<space>OVERWRITE or APPEND
where: C:\MYMSDS\DATA\DATAFILE.TXT is the full pathname of the file containing the spectra, and one of the words OVERWRITE or APPEND is the instruction as to how the spectra are added to the Spec List Window. To enable switching the focus back from the MS Search Program, the second line containing main window handle numbers of the data system application may be added (see Figure 41 for details).
This second locator file is deleted each time the NIST MS Search Program is brought into focus and spectra are imported. Figure 41 illustrates the implementation. The upper part of each box contains a file name; the bottom part is the contents of the file. Black arrows show logical connections between the files.


|
NIST MS Search (nistms$.exe) Command Line Options |
|
|
Option |
How it works |
|
/INSTRUMENT |
Normal search. Imports and optionally searches spectra (see Figure 41 for details on the implementation), activates MS Search Window, activates switch back button. |
|
/INSTRUMENT /PAR=2 |
Background search. Imports and optionally searches spectra (see Figure 41 for details on the implementation), does not activate MS Search Window (in fact, it is disabled during the search), writes Hit Lists into file SRCRESLT.TXT; when the search is complete creates file SRCREADY.TXT as a signal that SRCRESLT.TXT is ready. Both files are created in the MS Search work directory. Number of Hits is determined by the “Number of Hits to Print” in the Automation tab (Figure 30). |
|
/PAR=4 |
Start logging. Appends Hit Lists of Spectrum and Structure Searches to the file NISTLOG.TXT located in the MS Search work directory. Number of Hits is determined by the “Number of Hits to Print” in the Automation tab (Figure 30). |
|
/PAR=8 |
End logging. Stops appending Hit Lists to the file NISTLOG.TXT. |
|
/MOL=”<full pathname>” |
Imports chemical structure from a Molfile or SDfile. If SDfile contains more than one Molfile, only the first one will be imported. See section USE WITH THIRD-PARTY DRAWING PROGRAMS for details. |
SEE APPENDIX 1 for instructions on the Creation of AUTOIMP.MSD.
Use with Third-Party Drawing Programs
There is a Structure Search command. If a structure is in the Spec List pane of the Lib. Search tab or a structure is associated with a spectrum, this structure can be searched to retrieve spectra of all compounds with similar structures by selecting the Structure Search button (second button from the left) with the structure highlighted in the Spec List pane or by use of the Right Mouse button (RMB) menu with the Mouse pointer on the spectrum containing a structure.
![]()
When the NIST MS Search Program saves a structure to the Windows Clipboard using the Right Mouse button menu’s command Copy Structure to Clipboard, the structure is copied to the Windows Clipboard and a file in Molfile format named Clipboard.MOL is written to the NIST MS Search Program’s directory (by default, C:\NIST11\MSSearch). Other programs can either use the structure as it appears on the Windows Clipboard or this file for structure importation.
Structures saved to the Windows Clipboard may be accepted in the MS Search Program’s Librarian Spectrum Information dialog box if its Clipboard Struct button is not grayed. Some drawing programs need to be told how to save a structure in the Windows Clipboard. For example, in ISIS/Draw (MDL Information Systems, Inc.), it is necessary to select Copy Mol/Rxnfile to the Clipboard check box in Options/Settings/General tab of that program.
If an application is associated with files that have an.MOL extension, then the structure may be sent to that application by selecting in the RMB menu Send To and Default Structure Editor.
If a spectrum containing a chemical structure is selected, the RMB menu’s command Send to will result in a Send to menu that can have up to three additional structure editor items. These Send to menu items are a function of the first three lines in a text file AUTOIMP.STR located in the NIST MS Search Program’s working directory (the WorkDir32 item in [NISTMS] section of the WIN.INI file). Each line should contain three double‑quoted strings separated by spaces:
1) A program name as it appears in the Send to menu
2) Program executable and path
3) Program startup parameters. %1 will be replaced by the mol file name.
The following is an example of the contents of an AUTOIMP.STR file:
"ChemSite" "C:\MolSuite\ChemSite\ChemSite.exe" "%1"
"ISIS Draw" "D:\ISIS Draw\IDraw32.exe" "%1"
"ACD/ChemSketch" "C:\acd800\chemsk.exe" "%1"
The last line of the above example results in ACD lab’s ChemSketch starting with the contents of clipboard.mol being imported when ChemSketch opens.
When a program that is to be interactive with the NIST MS Search Program is installed, it must UPDATE or CREATE the AUTOIMP.STR file, or the AUTOIMP.STR file must be edited by the user before the third-party program will appear on and function from the Send to menu.
SEE APPENDIX 1 for instructions on the creation of AUTOIMP.STR.
A third-party program can call the NIST MS Search Program by issuing the command:
C:\NIST14\MSSEARCH\NISTMS$.EXE /MOL=<molfile>
assuming the NIST MS Search Program is in the C:\NIST11\MSSEARCH directory; if it is in some other directory, make the appropriate substitution (see Location of the NIST MS Search Program above)
where <molfile> is a full pathname of the MOL file. If there is a space in the name (e.g., example c:\program files\a.mol), the name should be double quoted:
C:\NIST14\MSSEARCH\NISTMS$.EXE /MOL="c:\program files\a.mol"
A third-party program can obtain the path to NISTMS$.EXE from Path32 in the [NISTMS] section of WIN.INI (see Location of the NIST MS Search Program in the USE WITH INSTRUMENT DATA SYSTEMS section above).
User Libraries
The NIST MS Search Program can create user libraries to search against. Up to 127 MS libraries, including the libraries from NIST 14 MS Library are allowed The user libraries may be searched in conjunction with the NIST/EPA/NIH Mass Spectral Main and/or Replicates Libraries, or they may be searched separately. The Program allows for the searching of combinations of libraries in any order. The inclusion of libraries and their order of search can be set from the Libraries or Options tab of the various Search dialog boxes (see Figure 28 and Figure 32 as examples).
User libraries are maintained as subdirectories under the MS Search Program’s working directory. The name given to the user library is the name of the subdirectory. This subdirectory will contain a number of library files and indices constituting the user library. The spectra and compound information are put in these files through the Librarian tab or by using Lib2NIST library utility.
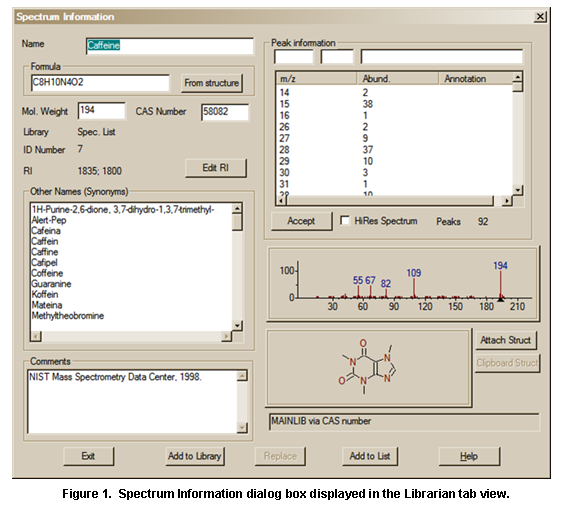 |
The spectra and compound information can originate in a text file in the NIST Text file format described in the NIST TEXT FORMAT of INDIVIDUAL SPECTRA section of this manual or directly imported from AMDIS. Spectra can be edited using with the Edit Spectrum button in the Librarian tab Tool Bar Figure 43 This will call up the Spectrum Information dialog box Figure 42. This dialog box can be used to add structures to user spectra from the clipboard, add identification information such as CAS number or synonyms and to add comments. In addition the RI value for the spectrum can be either added of edited.
In addition, a new spectrum can be entered using the New spectrum button in the Librarian tab Tool Bar (Figure 43).
The User libraries are easy to create and maintain. It may be desirable to create them using specific sets of spectra from the NIST/EPA/NIH Main or Replicates Library. Any spectrum in the Spec List can be added to a user library. Any spectrum in the Hit List of the Lib. Search tab, Other Search tab, or Names tab, a Plot pane, Text Info pane, or Subtraction pane in the Compare pane of the Lib. Search tab or the Compare tab can be copied to the Spec List pane by selecting Spec List on the Send to menu from the Right Mouse button menu. The size of a user library is limited to 65,535 spectra.
 |
The MS Search Program can read user‑submitted spectra in the JCAMP, U.S. EPA, or NIST Text file formats as well as formats of files submitted by various instrument manufacturers’ mass spectrometry data systems. The manufacturer of a specific mass spectrometer’s data system should be checked with to see if there is file compatibility.
Instructions on building user libraries are included in the Librarian Help screens. They also include the information necessary to add structures to user library spectra.
The “Tags in the “Comments” field of a user library spectrum can be used. A Tag is the name of a Field heading that will be displayed with the Text Information of a user library or Spec List spectrum and can optionally appear in the line below the Plot of the spectrum. Text is designated as a Tag in the “Comments” field of a user library or Spec List spectrum if it consists of the Field title (a line of characters without spaces) followed by an = (equals sign). Following the = is the contents of Field. This can be a line of characters without spaces, a number, or a text string enclosed in quotation marks (" "). For example, the following may be entered in the “Comments” field of the Spectrum Information dialog box of a Spec List or a user library spectrum:
|
Structures were prepared using SoftShell's ChemWindow Version 6.0 Contributor="University of the Pacific Mass Spectrometry Facility" Instrument="Agilent 5975 Inert XL MSD w/7890 GC" Tune="Standard Spec w/PFTBA" ScanRange=35-400 GC_column="30 m x 250 µm with 0.3 µm film thickness of BD-5" OvenTemp="50 C 5 min to 250 C @ 5 C/min" Synthesized="Matt Curtis". |
As shown in Figure 45, the information preceding each equals sign (=) will be displayed as a Field title in the Text Info pane of the spectrum and can optionally be displayed with the Plot pane of the spectrum. The information following the equals sign (=) will appear following the Field header. Entry of Field contents (string) must conform to the rules above. In order for this display to occur, the Field titles must be registered in the Display comment field options dialog box shown in Figure 44 (called from the Comment field options selection of the View menu. When the Tags have been entered into the Comment field options dialog box, they will not be displayed in the “Comments” field of the Text Info pane.
When a search is performed of a user library that contains Tags (Field titles), it is possible to constrain that search based on the Tag (Field title) and the partial or whole contents of the Tag (Field title). More specific details on the use of Tags as Constraints are in MS Search Program’s Help file.
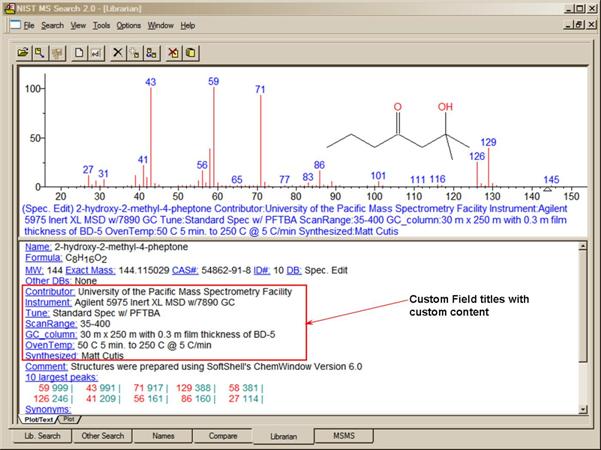
NIST Text Format of Individual Spectra
The NIST Text file format is simple. A file can contain as many spectra as wanted. Each spectrum must start with the Field title “Name:”. There must be something in this field in order for the spectrum to be imported (i.e., the data file name and the scan number of the spectrum’s source). The next required Field title is “Num Peaks:”. The contents of this field must be the number of mass/intensity pairs that make up the spectrum. Optional fields with the titles of “Comments:”, “Formula:”, “MW:”, “Synonym:”, and “CAS#:” can be between the “Name:” and “Num Peaks:” fields.
When a spectrum is saved to a text file from the NIST/EPA/NIH Main or Replicates Library, it will also contain fields with the names “NIST#:” and “DB#:”. The field with the name “NIST#:” is on the same line as the “CAS#:” field and separated by a semicolon (;). “DB#” is same as the “ID#” displayed in the Spectrum Text Window. The mass/intensity list begins on the line following the line with the Field title “Num Peaks:”. The lines of the mass/intensity list have no Field title.
When a file is created with all the allowable fields, each Field title should be on a separate line. Use the following format. The format for each spectrum in a text file usable by the Program should be as follows:
Line 1: NAME: Molecule (Required, up to 511 characters)
Line 2: COMMENT: Run 23, 8/8/88 (Optional, up to 1023 characters)
Line 3: FORMULA: C6H6 (Optional, up to 23 characters)
Line 4: MW: 78 (Optional)
If the spectrum submitted to Library spectrum Search contains a CAS registry number with its appropriate prefix (CAS: ), the display of the spectrum in the Plot, Compare, and Structure Windows will have the structure of the compound with that CAS registry number if present in the NIST/EPA/NIH Main Library.
Line 5: CAS: 71-43-2 (Optional)
Line 6: SYNONYM: Chemical name synonym (Optional, may be repeated)
The actual mass spectral data (number of peaks in the spectrum) may begin on the next line. It does not actually matter what line this is, as long as it precedes the line that starts the mass/intensity pair data. The VERY NEXT line and subsequent lines MUST contain the paired mass/intensity values.
Line 7: Num Peaks: 18 (This prefix and the exact number of mass/intensity pairs is Required.)
Line 8: 26 430; 27 340; 28 40; 37 480; 38 611; 39 1411; 49 300; 50 1792;
Line 9: 51 2052; 52 1962; 63 340; 73 160; 74 480; 75 180;
…
Line n: 76 721; 77 1401; 78 9806; 79 651;
The peaks need not be normalized, and the masses need not be ordered. The exact spacing and delimiters used for the mass/intensity pairs are unimportant. For example, the peaks above could equally be presented as:
(26,430),(27,340),(28,40),(37,480),(38,611),(39,1411),
(49,300),(75,180),(50,1792),(51,2052),(52,1962),(63,340),
(73,160),(74,480),(75,180),(76,721),(77,1401),(78,9806),(79,651)
or each mass intensity pair could be on an individual line. The following characters are accepted as delimiters (except “|”): |space|tab|,|;|:|(|)|[|]|}|
The file can be given any valid file name; however, it is best to use the extension “MSP”. This is the recognized default extension in the MS Search Program when user-library spectra are to be imported.
Several examples of NIST text format mass spectral files are installed together with the MS Search Program, namely SAMPLIB.MSP, UNKNOWN.MSP, and USERDEMO.MSP.
Import and Export of Retention Indices with a Spectrum
Retention index (RI) format was designed to provide compatibility with existing EI libraries and import of a mass spectrum into NIST MS Search.
In general, each RI value must have GC Column Type associated with it. Currently, there are 3 specific Column Types (1-3 in the table below) and 2 other (4-5) column types. RI of these two column type may be compared with any selected specific Column Type:
|
|
Column Type |
Accepted Tags (case insensitive) |
|
1 |
Semi-standard non-polar |
s, db5, semi, SemiStandardNonPolar, ssnp, SemiStdNP |
|
2 |
Standard non-polar |
n, db1, nonpolar, snp, standard, StandardNonPolar, StdNP |
|
3 |
Standard polar |
p, pol, polar, sp, StandardPolar, stdp, StdP, StdPolar |
|
4 |
Any (n-alkane standard) |
a, all, AllColumnTypes, AnyColumnType, std, Any |
|
5 |
Unspecified |
u, un, uns, unspecified, User |
When importing or adding a spectrum with RI of Unspecified column type to a library, there is an option of changing RI column type to any of the three specific Column Types.
RI Export with a Spectrum in a MSP or JCAMP file
A compound may have up to 3 RI values of different specific Column Types or a single RI of any of other column type. Actually used by NIST MS Search RI values are exported into a single line in a MSP file in the following format with Accepted Tags of up to 3 different specified Column types:
Retention_index: SemiStdNP=654/11/226 StdNP=654/7/286 StdPolar=957/17/131
or
Retention_index: Any=1000
or
Retention_index: User=2059
Slash-delimited numerical values following Accepted Tags are
Experimental RI median/deviation/number of datapoints
In NIST MS Search Spectrum Text Information window, RI values of the first example (Benzene) are displayed as
Experimental RI median±deviation (#data)
Semi-standard non-polar: 654±11 (226)
Standard non-polar: 654±7 (286)
Polar: 957±17 (131)
Other Unspecified RI values possibly present in the Comment (see the next section) are left unchanged in the exported spectrum.
Single RI value[5], if available, is exported into a JCAMP file as
##$RETENTION INDEX=654
RI Import with a Spectrum in a MSP or JCAMP file
Legacy MSP file formats allow saving several possibly different RI values of Unspecified column type, which may contradict each other. NIST MS Search picks up only one of them according to the priority rules described below.
If the last out of all lines starting with Retention_index: or RI: or Synon: $:29 in a MSP file has format described in the previous section with possibly different Accepted Tags, then contents of this line is used as input.
Otherwise, there may be up to 4 untagged RI value locations/formats of Unspecified column type. They are described in the following table (n is the RI value):
|
Priority |
Format |
Location |
|
Highest |
|RI:n |
Last occurrence in comment |
|
|
RI=n |
Last occurrence in comment |
|
|
RI:n |
Last occurrence in comment |
|
Lowest |
Retention_index:n or Retention_index=n or RI:n or RI=n or Synon: $:29n |
Last occurrence in MSP file |
RI value imported from a JCAMP file always has Unspecified column type.
APPENDIX 1: Creating AUTOIMP Files Used in NIST MS Search
It may be necessary to create AUTOIMP.MSD as the primary locator file used for the importation of spectra from third-party mass spectral data systems and/or AUTOIMP.STR for interaction between the MS Search Program and third-party drawing programs. These files may be created using Notepad with certain versions of Microsoft Windows.
If Microsoft Notepad is used, the default extension is .TXT (Text Document) even though AUTOIMP.MSD or AUTOIMP.STR has been entered in the “File name:” field. The default settings for Windows Explorer will not show this .TXT extension. The file will not work unless the .TXT extension is removed. The file can be renamed by highlighting the file in Windows Explorer and selecting Rename from the File menu.
When saving the file using Save As option, this problem may be completely avoided by selecting All Files in the Save as type drop-down list box (Figure 46). In this case, the file will be saved with the extension provided in the “File name:” field (see the snapshot of the Save As dialog box below).
When saving the file using the Notepad Save As option, also make sure the ANSI Encoding is selected (in some versions of Windows, the Encoding option is not present).
After File name, Save as type, and Encoding are set correctly, press the Save button.
|
APPENDIX 2: Using the NIST MS Search Program with ChemStation
The NIST MS Search Program has been supported through several versions of the HP/Agilent Technologies ChemStation. In the recent versions of the ChemStation for GC/MS, both the NIST Output and the AMDIS options will appear on the Spectrum menu of the Data Analysis software when using the Enhanced Quantitation, Standard Quantitation, or Gasoline mode; and on the Lib. menu when using the Environmental mode if the appropriate macros have been installed. The NIST/EPA/NIH MS Library (NIST 14) setup will install the two required macros along with additional macros that will allow for greater flexibility with the ChemStation software.
It is possible to use the ChemStation double-click option on a displayed spectrum to send that spectrum to the NIST MS Search Program’s Spec List Window, bringing the MS Search Program into focus. If the appropriate options (including Automation, see Figure 27 and Figure 30) have been set in the NIST MS Search Program then, after the spectrum is automatically prepended to the Spec List Window, it will be searched.
If using the Enhanced Quantitation mode on any version of ChemStation, the setup will automatically add four new options to the Spectrum menu (PBM Quick Search, NIST Search, Multiple Lib PBM Search, and Set Default Search Engine). Each of these menu options is explained below. If these options are to be added to other modes of the ChemStation software, see the MENU_MOD document file on the NIST MS Search Program installation CD ROM in the \support.cs directory. This document is also available after installing NIST 14 MS Library by selecting ChemStation Menu Modification from the NIST Mass Spectral Database program folder.
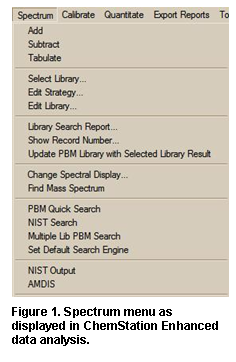 Some older versions of ChemStation
will have ASCII Output as opposed to NIST Output, the Spectrum
menu, or the Lib. menu found on the Environmental mode. Selecting
either the NIST Output or ASCII Output results in the display of
the Options for NIST Library Search dialog box with six radio button
options and OK and Cancel buttons. The Macro NISTINT.MAC must be
installed in the HPCHEM\MSEXE or MSDCHEM\MSEXE directory for these menu
selections to be displayed.
Some older versions of ChemStation
will have ASCII Output as opposed to NIST Output, the Spectrum
menu, or the Lib. menu found on the Environmental mode. Selecting
either the NIST Output or ASCII Output results in the display of
the Options for NIST Library Search dialog box with six radio button
options and OK and Cancel buttons. The Macro NISTINT.MAC must be
installed in the HPCHEM\MSEXE or MSDCHEM\MSEXE directory for these menu
selections to be displayed.
When all the necessary macros have been installed in the ChemStation’s MSEXE directory and the modifications have been made to the ChemStation macros, the following options will be available from the ChemStation Spectrum menu (Figure 47):
The PBM Quick Search (PBMSrch.mac) performs the normal ChemStation PBM search of the currently displayed spectrum.
The NIST Search (NISTSrch.mac) passes the currently displayed mass spectrum to the NIST MS Search Program. The spectrum will be prepended to the NIST MS Search Program’s Spec List Window. If the NIST MS Search Program is configured to search automatically, then the search will be started when the spectrum is passed to the NIST MS Search Program. Otherwise, double clicking on any spectrum in the Spec List Window located in the Lib. Search tab to start a search.
The Multiple Lib PBM Search (LS.mac) performs a ChemStation PBM Search using up to three different libraries (all of which have to be specified using the dialog box displayed by selecting the Select Library option on the Search menu). The top 20 Hits from each library will be displayed in order of decreasing match quality.
Set Default Search Engine (SWITCH.MAC) displays a dialog box that allows the selection of the default library search to be evoked when double clicking the RMB on the currently displayed spectrum. The NIST MS Search is the default until another search type is set as the default.
NIST Output (NISTINT.mac) displays a self‑explanatory dialog box that allows for the use of the NIST MS Search Program in a number of different ways and the copying of ChemStation user libraries as NIST MS Search Program user libraries.
AMDIS (AMDIS.mac) loads AMDIS with the data file currently being viewed with the ChemStation Data Analysis directly into AMDIS for analysis. Once in AMDIS, select Run to perform the analysis.
The Lib2NIST converter program will be installed along with the NIST MS Search Program. This program allows mass spectral libraries that have been created using the internal utilities of the ChemStation software (including structures) and other mass spectral libraries to be copied to the NIST user libraries format that can be employed by the NIST MS Search Program. This utility is evoked as an option from the Options for NIST Library Search dialog box displayed by selecting the Copy ChemStation Lib to NIST Format radio button and then selecting the OK button or by selecting the Lib2NIST icon in the NIST Mass Spectral Database program folder.
An additional macro is found on the NIST MS Search Program’s installation disk, AUTONIST.MAC. This macro is to be incorporated into automated sequences. It automatically selects spectra based on the autointegration of the current data file and switches to the NIST MS Search Program where the spectra are automatically imported into the Spec List Window. If the proper settings have been made in the Program, these spectra are searched and the focus returned to the HP MS software. These features can only be utilized with G1034C, Version C.3.00.00 or later of ChemStation.
Additional Information Regarding Modes Other Than Enhanced Quantitation
During the installation of the NIST/EPA/NIH MS Library (NIST 14), if the NIST MS Search Program has been selected as the ChemStation’s default search engine, each time a spectrum is double clicked, the displayed spectrum, will be prepended in the NIST MS Search Program’s Spec List Window. The NIST MS Search Program will be brought into focus regardless of the data analysis mode (Enhanced Quantitation, Standard Quantitation, Gasoline, or Environmental). One of the other above-referenced search methods can be changed to by selecting the ChemStation Default Search Mode icon in the NIST Mass Spectral Database program folder.
MODIFICATION OF MACROS IS NOT NECESSARY TO SWITCH FROM THE NIST MS SEARCH PROGRAM AS THE DEFAULT SEARCH METHOD FOR THE DOUBLE-CLICK FEATURE OF THE ChemStation.
HP_NIST MACRO (NISTINT.MAC)
 When NIST Output is selected
from the Spectrum menu of the MS ChemStation or ASCII Output from
the Lib menu of the Environmental software, the Options For NIST
Library Search dialog box is displayed (Figure 48). This dialog box consists of six options and OK and Cancel
buttons. Each option has a radio button next to it. Use the Mouse
pointer and the left Mouse button (LMB) on the radio button to select an
option. Selecting the OK button results in execution of the option.
When NIST Output is selected
from the Spectrum menu of the MS ChemStation or ASCII Output from
the Lib menu of the Environmental software, the Options For NIST
Library Search dialog box is displayed (Figure 48). This dialog box consists of six options and OK and Cancel
buttons. Each option has a radio button next to it. Use the Mouse
pointer and the left Mouse button (LMB) on the radio button to select an
option. Selecting the OK button results in execution of the option.
Items in the Options For NIST Library Search dialog box are selected by putting the Mouse pointer on the appropriate radio button and clicking the LMB. Then select the OK button to execute the desired option. If any of the first three options is executed, the dialog box will be redisplayed.
Each time one of the top four menu selections is executed, the file HPNIST.TXT is created in the ChemStation HPCHEM or MSCHEM directory. This file contains the name and location of the text file that contains the saved spectra in the NIST format. The HPNIST.TXT file is deleted every time the NIST MS Search Program is brought into focus and spectra are automatically imported into the top of the NIST MS Search Program’s Spec List Window.
At the same time the HPNIST.TXT, the second locator file, is created, a text file named TEMPSPEC.MSP containing one or more spectra is created or appended. The text file is located in the ChemStation directory HPCHEM or MSDCHEM. This text file will remain there until it is overwritten or removed manually.
When the NIST MS Search Program comes into focus, the spectra that have been saved in the ChemStation software are automatically copied to the Spec List Window. If spectra are already in the Spec List Window, a prompt to Prepend or Overwrite the current contents will be displayed. If Prepend is selected, the new spectra are added to the top of the Spec List Window (prepended). If Cancel is selected, the automatic import will be aborted. However, the HPNIST.TXT file may be deleted even in the case of an abort. If the options of Automatic Search On and Return Focus to Caller upon Completion have been selected in the Automation dialog box, selecting Cancel returns the focus to the ChemStation software.
Menu “Options for NIST Library Search”
The Create Integrated Spectra File option runs the ChemStation’s Autointegrate routine on the displayed chromatogram and creates the text file containing the spectra that represent the apex of each chromatographic peak.
The Create File with Spectrum in X option creates a text file that contains the spectrum that is in X of the ChemStation Stack display. This is where background-subtracted and averaged spectra will be found. This is the currently displayed spectrum.
The Append File with Spectrum in X option appends the spectrum in X of the Stack display to the existing text file. If a text file does not exist, one will be created.
The Create\Search Integd. Spect. File option creates a text file that contains spectra representing the tops of each chromatographic peak and either starts or switches to the NIST MS Search Program where the spectra are automatically imported into the Spec List Window. If Automatic Search On in the NIST MS Search Program’s Automation dialog box has been selected, each spectrum will be searched. The results are optionally printed, and the results of each search are stored in the Hit List History. If Return Focus to Caller upon Completion has been selected in the Automation dialog box, then after the last spectrum is searched and printed, the focus will be returned to the ChemStation software. If spectra are already in the NIST MS Search Program’s Spec List Window, the Append/Overwrite/Cancel query will be displayed.
The Toggle to NIST Library Program option either starts the NIST MS Search Program or switches to it if it is already running. The ChemStation software can be redisplayed by holding down the <Alt> key and pressing the <Tab> key, or the NIST Program’s Switch To Caller button can be selected. The NIST MS Search Program can be brought back into focus by using the <Alt><Tab> command or by selecting the Toggle option from the Options For NIST Library Search dialog box.
The Copy ChemStation Lib to NIST Format option is described in APPENDIX 3.
AUTONIST Macro
This is a non-interactive macro that can be executed using the command line instruction:
MACRO “AUTONIST.MAC”, go
or from within the Method. Every Method directory contains a file DEUSER.MAC. Edit this file to contain the string:
MACRO “AUTONIST.MAC”,go.
This File is in the \SUPPORT.HP directory of the CD ROM and must be copied to the \HPCHEM\MSEXE directory.
When the Method is run, the AUTONIST Macro will execute. The chromatogram is autointegrated, and the spectra representing the top of each chromatographic peak are stored in a text file. The focus is then switched to the NIST MS Search Program. The spectra are imported into the Program’s Spec List Window. If spectra are already in the Spec List Window, they will be overwritten.
If Automatic Search On and Return Focus to Caller upon Completion have been selected in the NIST MS Search Program’s Automation dialog box, the spectra will automatically be searched. After the last spectrum is searched, the focus is returned to the HP MS software. In order for this macro to function properly, it will be necessary to have selected the above two options in the Automation dialog box.
APPENDIX 3: Copy ChemStation and Other Database to NIST User Library Format
This is a separate program called by selecting the Lib2NIST Converter icon on the NIST Mass Spectral Database program folder. This program allows mass spectral libraries in a variety of formats (along with structures or structures in separate files) to be selected and copied to the NIST user library format. For all 32-bit Windows (Windows NT, Windows 98/Me/2000/XP/Vista/Windows 7, 8, 8.1) and Windows 64-bit platforms, the Lib2NIST converter program replaces the HP2NIST program that was provided with previous versions of the NIST MS Search Program. The use of the HP2NIST program is described at the end of this section. Another feature of the Lib2NIST program is the ability to take user libraries that were developed by using versions 1.5 through 1.7 of the NIST MS Search Program and copy them into a format fully compatible with the current version of the NIST MS Search Program.
Select the library to be copied and then click on the Open button (Figure 49). The drop-down arrow will also allow for selecting from a variety of JCAMP formats. More than a single library can be selected in this dialog box by using the standard Windows multiple file selection techniques.
 The selected libraries will now be
listed in the Input Libraries or Text Files pane of the Program’s
display (Figure 50).
The selected libraries will now be
listed in the Input Libraries or Text Files pane of the Program’s
display (Figure 50).
The NIST Library and Output locations can be changed by selecting the appropriate button located to the right of the two fields. This will result in the display of the Browse for Folder dialog box (Figure 51).
Select the desired directory (folder) and then click on the OK button to change the location.
The Define Subset button will display a dialog box that allows for the choice of a range of input spectra ID or CAS numbers. The Use subset check box is grayed, unless an entry has been made in the Define Subset dialog box (Figure 52).
After making any desired entries in the Define Subset dialog box, select the OK button to return to the Program’s main display.
Figure 52. Define Subset dialog box.
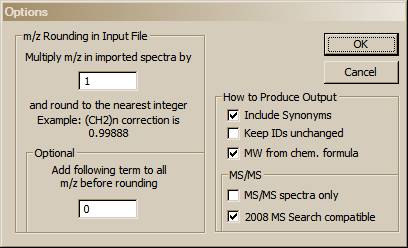
Figure 53. Options dialog box display.
The Options button on the Program’s display will display the dialog box shown above in Figure 53. This allows for the entry of mass defect corrections to be applied to m/z values of imported spectra and whether or not to: 1) include synonyms from the source file, 2) retain the ID numbers (or sequence numbers) from the source library or file[6], 3) calculate molecular weights (nominal mass) from formula, and 4) create or convert MS/MS and/or In-source libraries[7].
Select the libraries to be converted in the Input Libraries or Text Files pane of the Program’s display, make sure the Output Format is selected correctly, and then click on the Convert button. The selected libraries will then be converted and placed in the specified output directory. Advanced features of Lib2NIST are explained in the file CMDLINE.pdf installed with the Lib2NIST. Transliteration rules of extended ASCII characters are listed in the ASCII text file HPTRANS.TBL and may be modified in this file by the user.
APPENDIX 4: Using the NIST MS Search Program with Thermo Fisher Corporation Xcalibur Software
The Thermo Fisher Xcalibur software uses the NIST MS Dynamic Library (dll) as a library search engine. A copy of the NIST MS Search Program and AMDIS may be provided with a copy of the Xcalibur software. The NIST/EPA/NIH Mass Spectral Library may be optionally provided by the Thermo Fisher Corporation.
To add the NIST/EPA/NIH Mass Spectral Libraries to the Xcalibur software, run the NIST 14 MS Library setup. If it locates the following NIST MS Search Program and the AMDIS directories:
C:\Program Files\NISTMS\MSSEARCH
C:\Program Files\NISTMS\AMDIS32
then let the setup install NIST 14 in these directories. The NIST 14 MS Library will then be properly installed along with any necessary program updates.
More recent versions of the Xcalibur software detect and use NIST MS Search installed in another directory (for example, c:\nist11\mssearch) and the NIST/EPA/NIH Mass Spectral Library using means explained in the USE WITH INSTRUMENT DATA SYSTEMS section of this manual.
Exporting MS/MS spectra to NIST MS Search transfers both the mass spectrum and the precursor m/z values.
Figure 54. Exporting a spectrum to NIST MS Search.
APPENDIX 5: Search Algorithms
There are two general ways that the NIST MS Search Program can retrieve library spectra that resemble the submitted spectrum. They are the Identity Search and the Similarity Search. An Identity Search is designed to find exact matches of the compound that produced the submitted spectrum and therefore presumes that the unknown compound is represented in the reference library. Only experimental variability prevents a perfect match. The Similarity Search is optimized to find similar compounds and is intended for use when a compound cannot be identified by the Identity Search (it is probably not in the library).
Screening
For the sole purpose of achieving rapid retrieval rates before actually comparing spectra, modern search algorithms first identify a subset of library spectra with important features in common with the unknown spectrum. While this can vastly reduce search times, it can also screen out the correct spectrum. When this occurs, the correct retrieval cannot appear in the Hit List. Because of unavoidable blind spots in simple algorithms, even a closely matching spectrum can be excluded. This is like “throwing out the baby with the bathwater”. This is probably the most serious failure of any mass spectral search system.
A variety of filtering algorithms for Identity searching were tested in an effort to avoid throwing away correct matches without sacrificing performance. These algorithms were tested using the 12,592 spectra that comprised an older version of the NIST/EPA/NIH Selected Replicates Library. The best performing algorithm used “a ranked peaks in common” logic similar to that incorporated in existing data systems. This finds library spectra with the largest number of peaks in common with the unknown spectrum consistent with a required minimum number of identified spectra. Tests showed this minimum number to be about 50. This procedure retrieved 95% of the matching compounds (5% of matching compounds were lost). By scaling peaks by their m/z values, a 98% success rate was achieved at the same search speed. This is the screening logic of the Quick Identity Search option. By combining results using several screening criteria, a 99.4% success rate was achieved with a modest reduction in search speed. At this level, virtually all correct matches that were screened out were very dissimilar to the unknown spectrum and would have produced low Match Factors. Both peak scaling and merging of multiple screening results are used in the Normal Identity Search. A more detailed description follows:
Peak Scaling: The determination of the largest peaks in a spectrum was made after first multiplying the abundance of each peak by the square of its m/z value. The most intense peak in the scaled unknown spectrum is compared against the eight most intense peaks in scaled library spectra. The second most intense peak in the scaled unknown spectrum is then compared against the nine most intense peaks in the library spectra. This is repeated in decreasing order of intensity of peaks in the scaled spectrum of the unknown until the eighth most intense peaks are compared against the 16 most intense peaks in the scaled library spectra. This is the only screening procedure applied in the Quick Identity Search.
Merged Subsets: Since the use of any single set of peak specifications was found to fail for certain classes of spectra, results of multiple sets of peak specifications were merged to reduce this problem. After extensive optimization studies, four separate peak specifications were selected. The first of these specifications is described above for the Quick Search. The others were:
1) The fourteen largest peaks in the scaled unknown spectrum were matched against the fourteen largest peaks in the scaled library spectra.
2) The six largest peaks in the original (non-scaled) spectrum were matched against the six largest peaks in the original library spectra.
3) The five largest peaks along with the “maximum mass” peak in both the unknown and library spectra were matched.
The screening algorithms for the Similarity Search are similar to the Normal Identity Search except that scaling and maximum mass peaks are not used. When neutral loss peaks are used in the Hybrid and Neutral Loss Search, up to five neutral loss peaks within 64 m/z of the molecular ion are used in place of conventional peaks. For neutral loss peaks, abundances in library spectra are required to be within a factor of four of the abundances of corresponding unknown spectra (peak ranking and scaling are not used).
Search
A mass spectrum can be represented as a row vector composed of the ordered peak intensities. It can also be considered to represent a single point in a multidimensional hyperspace defined by the m/z variables. Each of the intensities in the row vector represents the value of the coordinate of the spectral point along the individual mass axis in this hyperspace. If two spectra being compared are identical with respect to all the mass intensity pairs, their point representations in this hyperspace will coincide. If these spectra are very similar, their point representations will be close to one another. The Match Factor, which provides a sense of spectral similarity, may be regarded as the inverse of distance of the two point representations when each spectral vector has unit length.
The dot-product mass spectral search algorithm, which uses the cosine of the angle between the unknown and library spectral vectors, has been optimized by scaling peaks using the square root of their abundance. For the Identity Search, peaks were weighted by the square of their m/z value and a second term was added that compares ratios of adjacent peaks in library and unknown spectra. Its contribution was weighted so that it increased in importance as the proportion of common peaks increases. The only difference in the application of this algorithm to the Identity Search and Similarity Search is that m/z weighting is used only in the former. This defines the search algorithm used for both the Similarity and the Identity Searches in the Program.
For additional information, see Stein, S.E. “Optimization and Testing of Mass Spectral Library Search Algorithms for Compound Identification” J. Am. Soc. Mass Spectrom. 1994, 5, 859–865.
APPENDIX 6: Syntax for Some Constraints in Searches
Syntax of Name Fragments Constraint
Applies to: all searches
Max. length: 39 characters
First release: 2000
This “Name Constraint” field can contain up to eight name fragments.
The name fragments are searched in compound main chemical name and in other names (synonyms).
The following examples explain the conditions when the name constraint is satisfied (spaces around && and !! are optional):
"COCA && INE": at least one name contains both "COCA" and "INE"
"COCA && !! INE": at least one name contains "COCA" and doesn't contain "INE"
"COCA !! INE": at least one name contains "COCA"; no name contains "INE"
Syntax of Tags in Comment Constraint
Applies to: all searches
Max. length: 1023 characters
First release: 2006
All information in NIST Peptide libraries is represented by the Tag=value convention. Typically, the “Peptide spectrum comment” field may contain the following items separated by spaces:
Tag=string
Tag="string"
Tag=number
Spaces may be inside a string only if the string is in double quotes. In older libraries, an unquoted string may also have space(s) inside a pair of parentheses ( ). Spaces are not permitted inside any Tag or next to the equal sign.
“Comment” fields in older libraries may also contain Tag not accompanied by the equal sign and string.
All Tag, string, substring, and word comparisons described below ignore letter case.
Entering Tags in Comment Constraint
Items in the “Tags in Comment” constraint field should be separated by spaces and/or linefeed characters. Unlike spectra comments, spaces next to the equal sign (as well as >, <, >=, <=, and :) are allowed inside the items.
Spectra returned must satisfy all entered constraints.
Examples
|
|
Item |
Comments of the output spectra must have |
|
1 |
"consensus" |
A Tag or a space-delimited string consensus |
|
2 |
parent |
6 letter sequence parent anywhere in the Comment |
|
3 |
fullname=”r.lecvk.c/2” |
Exactly this Tag=string:: Fullname=R.LECVK.C/2 |
|
4 |
fullname=lec |
String following Fullname= contains sequence LEC |
|
5 |
Mods : ICAT_cl_hi |
String following Mods= contains word1) ICAT_cl_hi |
|
|
The following rules apply to strings that are numbers |
|
|
6 |
Diffmod=6.8 |
String following Diffmod= begins with number 6.8, for example, Diffmod=6.8/8 or Diffmod=6.8 |
|
7 |
Diffmod=5.1:6.8 |
String following Diffmod= begins with number between 5.1 and 6.8, for example, Diffmod=6.8/8 or Diffmod=6.8 |
|
8 |
Diffmod > 6.8 Diffmod >= 6.8 Diffmod < 6.8 Diffmod <= 6.8 |
Diffmod= followed by a number greater than 6.8 Diffmod= followed by a number not less than 6.8 Diffmod= followed by a number less than 6.8 Diffmod= followed by a number not greater than 6.8 |
|
1) word is a string delimited by any character but a letter, a digit, or an underscore “_”; If word contains spaces or equal sign it should be entered in quotes. |
||
To reverse the meaning of constraints 1–5 (that is, change to “spectra must not have”), enter caret ^ as the first letter:
|
1 |
^"consensus" |
Tag consensus is not present |
|
2 |
^parent |
Substring parent is not present |
|
3 |
fullname=^"r.lecvk.c/2" |
String tagged fullname is not exactly r.lecvk.c/2 |
|
4 |
fullname=^lec |
String tagged fullname does not have substring lec |
|
5 |
Mods : ^ICAT_cl_hi |
String tagged Mods has not word ICAT_cl_hi |
More formal description
|
|
Item |
Comments of the output spectra must have |
|
1 |
"Tag" |
Exactly this Tag is present in the comment, possibly not followed by the equal sign |
|
2 |
substring |
this substring anywhere in the comment |
|
3a |
Tag="string " |
exactly this string tagged with this Tag= |
|
3b |
Tag="" |
This Tag has no string or its string is “” |
|
4 |
Tag=substring |
this substring in the string tagged with this Tag= |
|
5a |
Tag : word |
exactly this word inside string tagged with this Tag= |
|
5b |
Tag : "word" |
exactly this word inside string tagged with this Tag= |
|
6 |
Tag=number |
tagged string = number or string begins with number |
|
7 |
Tag=number:number |
tagged string number inside the given range |
|
8a |
Tag >= number |
The number in the string less or equal to number |
|
8b |
Tag > number |
The number in the string less than the number |
|
8c |
Tag <= number |
The number in the string greater or equal to number |
|
8d |
Tag < number |
The number in the string greater than the number |
Notes.
· Inside an item, spaces are allowed next to signs =, >, <, >=, <=, or :. These signs as well as Tag and string or number or word may be located on separate lines.
· If a Tag created contains characters = or < or > or : then use double quotes to search for such a tag, for example, "<Tag>"="string ".
· substring in rows 2 and 4 may not contain characters < > = :.
· To search for a substring that may be interpreted as a number, for example, 1.23e5, use Tag==1.12e5 or Tag=="1.12e5" instead of Tag=1.12e5
· To reverse the meaning of constraints 1–2 (that is, change to “spectra must not have”), insert caret ^ in front of the "Tag" or substring
· To reverse the meaning of constraints 3–8 (that is, change to “spectra must not have”), insert caret ^ in front of the right hand side expression.
· To use caret ^ as the first character of a substring enter Tag == "^substring".
· In 3-8, Tag may be replaced with Tag? In this case the spectrum is accepted not only if the condition(s), described in the above table are satisfied but also if Tag is missing.
Example of a multi-item constraint: The following items
Replicate- Line 2, substring
RT >= 11.2- Line 8a, number is greater or equal
Sample = "yeast_Nature_CAM" - Line 3a, Tag=”string”
Datfile = 011599.dat- Line 4, Tag=substring
fit the following “Comment” field:
Replicate RT=11.220m Sample="yeast_Nature_CAM" Datfile="F011599.dat"
More formal description of reversed constraints
|
|
Item |
Comments of the output spectra must have |
|
1a |
^"Tag" |
Exactly this Tag is not present in the comment |
|
1b |
~"Tag" |
Some of the tags are not exactly this Tag |
|
2a |
^substring |
this substring is not anywhere in the comment |
|
2b |
~substring |
this substring is not anywhere in the comment |
|
3a |
^Tag="string" |
all tags but this Tag have exactly this string |
|
3b |
~Tag="string" |
some tags but not this Tag have exactly this string |
|
3c |
^Tag=^"string" |
all tags but this Tag haven’t exactly this string |
|
3d |
~Tag=^"string" |
some tags but not this Tag haven’t exactly this string |
|
3e |
Tag=^"string" |
this Tag has no string or its string differs from string |
|
3f |
Tag=^"" |
this Tag has a non-empty string |
|
3g |
all=^"string" |
Each of the tagged strings has not exactly this string |
|
3h |
Some="string" |
some tagged strings have exactly this string |
Items 4–8 from the previous section (1.2.2) may be given reversed meaning in the same way as 3a–3f.
Reserved expression all= is same as ^""= and ^=
Reserved expression some= is same as ~""= and ~=
To address a tag
named all or some put it in quotes, for example:
"all"=string "some"=string
Quoting tags allows searching for exact combination of tags, for example
“Single Tryptic_simple Parent”=358.07 or “Charge=2 Scan”=82
Tag?=^whatever is considered found if either Tag=^whatever has been found or Tag is not present.
Tags in Comment Constraint for MS/MS Text Information
Text information displayed for NIST 14 MS/MS Library mass spectra has special tagged text fields, for example:
|
MSn pathway: [M+H-H2O]+=>311.4 Precursor type: [311.4]+ Spectrum type: MS3 Precursor m/z: 375.2894, 311.4 Instrument type: IT/ion trap Instrument: Thermo Finnigan Elite Orbitrap Sample inlet: direct flow injection Ionization: ESI Collision gas: He Collision energy: 35% Ion mode: P Notes: Consensus spectrum; Acetonitrile/Water/Formic acid; Vial_ID=1741 InChIKey: KXGVEGMKQFWNSR-UHFFFAOYSA-N |
In MS Search v.2.2, Tags in Comment Constraint allows searching these tagged fields. Special tags are to be used for this purpose.
To enter such a special tag, type dollar sign ($) immediately followed by the tag displayed in the spectrum text information, omitting spaces. Slashes, dashes, underscores in the tag may be omitted or added. Tags differing only in CAS, CAS#, or CASNO at the end of the tag are equivalent. The special tag and search string are case-insensitive.
The syntax is almost the same as the one described above for Tags in Comment Constraint.
Below are several examples.
|
To search for |
Enter into the Tags in Comment Constraint |
|
precursor m/z in range* 374.9–375.4 |
$Precursor_m/z=374.9:375.4 |
|
Instrument type containing word IT |
$Instrument_type:IT |
|
the exact Instrument name |
$Instrument="Micromass quattro" |
|
spectra that have any Related CAS# entry |
$Related_CAS |
|
spectra that have no Collision energy entry |
^$CollisionEnergy |
* Note: MS3 spectrum with text information “Precursor m/z: 375.2894, 311.4” also will be found. That is, for MSN (N>2) spectra, the first precursor m/z (corresponding to MS2) will be found.
List of all acceptable special tags
|
$aux $charge $collision_energy $collision_gas $compound_type $cone_voltage $inchikey $in-source_voltage $instrument $instrument_type $ion_formula |
$ion_mode $ion_mw $ion_name $ionization $known_impurity $mass_range $maximum_intensity $msn_pathway $notes $peptide_mods $peptide_sequence
|
$precursor_m/z $precursor_type $pressure $related_cas# $retention_index $ri $salt $salt/mix_cas# $sample_inlet $special_fragmentation $spectrum_type
|
Special tags inside parentheses are considered identical:
($aux $notes) ($in-source_voltage $cone_voltage) ($retention_index $ri)
Some of these tags may be found in other NIST 14 EI Libraries, for example, $related_cas#, $inchikey.
See also MS Search Help topic "Spectrum Text Information"
MS/MS Instrument Type Constraint
This feature will only work with NIST 14 MS/MS Libraries or similarly formatted user libraries.
It affects only ms/ms spectra (that is, spectra which have precursor m/z value); therefore, it ignores all EI spectra, which means applying this constraint to a search of the EI Library does not affect the Hit List.
This constraint has 7 options (check boxes) that may be combined to produce a Hit List containing such spectra that each spectrum fits one of the selected Instrument Types.
The “Instrument type” field in a MS/MS mass spectrum text information is interpreted in the following way:
|
Instrument type check box |
“Instrument type” field in MS/MS mass spectrum |
|
Ion Trap |
“IT/ion trap” or “QQIT” or “QqLIT” |
|
Q-TOF |
“Q-TOF” |
|
Ion Trap + FT |
“IT-FT/ion trap with FTMS” |
|
HCD |
“HCD” |
|
Triple quadrupole |
“QqQ” |
|
Other |
any other than the above five instrument types |
|
Unspecified |
No instrument type specified |
Selecting all seven would select all mass spectra (the same as if the constraint is disabled).
The version of Lib2NIST included in NIST 14 or a later version is needed to produce such a library because the instrument type is saved in the NIST 14 MS/MS mass spectrum record as a new binary field; therefore, this constraint would not work with the NIST 08 MS/MS Library or even the NIST Peptide Libraries created with the previous version of Lib2NIST.
In case of NIST 11 or NIST 12 MS/MS libraries, Q-TOF check box would work as Q-TOF and HCD check boxes selected together, HCD and Ion Trap + FT would not find any spectra although 737 HCD spectra are present in each of NIST 11 and NIST 12 MS/MS library. However, HCD spectra in these libraries may be found by entering Tags in Comment constraint $Instrument_type=HCD or Name Fragment constraint $:06HCD
See Read Me Lib2NIST (Lib2NIST.TXT or Readme_Lib2NIST.txt) for more details on rebuilding MS/MS library.
APPENDIX 7: Contributing EI Mass Spectra to the NIST/EPA/NIH Mass Spectral Library
Users of the NIST MS Search Program are invited to submit mass spectra for inclusion in the NIST/EPA/NIH Mass Spectral Library. Descriptions of both ideal and minimum acceptable data are provided below. Please provide data that is as close to the ideal as convenient.
|
Characteristic |
Ideal |
Minimum Acceptable |
|
Substance |
Pure compounds (single GC component) of confidently known structure that are commercially important, occur in the environment, or are a natural product (including analytically useful derivatives) |
Any single chemical compound whose identity is confidently known |
|
Method |
Acquired by GC/MS at an ionization energy of 70 eV under conditions such that isotope ratios are not distorted and ions above the molecular ion are absent and processed using AMDIS or other deconvolution software |
Acquired at an electron ionization of not less than 20 eV under conditions such that isotope ratios are not grossly distorted and significant ions above the molecular ion are absent; significant background subtracted |
|
Requested Information: |
||
|
Contributor |
Contributor’s name and/or institution (as it is to appear with the spectrum) |
Contributor’s name and/or institution (as it is to appear with the spectrum) |
|
Chemical name |
Chemical name (preferably in Chemical Abstracts or IUPAC format) |
Chemical name (or structure) |
|
Chemical Abstracts Registry Number |
Chemical Abstracts Registry Number |
- |
|
Chemical structure |
Chemical structure (in MOL/SDF file format) |
Chemical structure (or name) |
|
Synonyms |
Alternative names (trade names, trivial names, etc. |
- |
|
Mass spectrum |
Mass spectrum (unit m/z resolution or better) with all observed nominal masses from no higher than m/z 12 to above the molecular ion with intensities of 0.1 % or greater and intensity in electronic format (preferably as an NIST/EPA/NIH user library). Ions at half-mass should be kept in the spectrum and rounded down to the nearest mass. |
Mass spectrum (unit mass resolution) with all observed nominal masses beginning no higher than one-third the molecular mass to above the molecular ion with intensities of 2.0%. Ions at half-mass should be kept in the spectrum and rounded down to the nearest mass. |
|
Instrument type |
Instrument type (magnetic sector, linear quadrupole, TOF, etc.) |
Instrument type (magnetic sector, linear quadrupole, TOF, etc.) |
|
Instrument model |
Instrument manufacturer and model name |
- |
|
Ion source temperature |
Ion source temperature (°C) |
- |
|
Inlet |
Inlet (GC, probe, etc.) and inlet temperature (of probe, reservoir, etc.) |
- |
|
Retention index |
Kovats Retention Index (with the commercial name of the GC phase or, if not available, whether the phase is polar or non-polar) |
- |
|
Impurities |
Note masses not belonging to the compound and their origin, if known. |
Note masses not belonging to the compound, if known. |
Submissions and inquiries may be directed to Anzor I. Mikaia, National Institute of Standards and Technology, 100 Bureau Drive Stop 8380, Gaithersburg, MD 20899-8380, USA. Anzor Mikaia, .
APPENDIX 8: Support Contacts
In our continuing commitment to quality, the NIST Mass Spectrometry Data Center is always looking to improve the quality of our Mass Spectral Libraries and programs for accessing them. Comments or questions about the quality of them should be made by contacting:
Adam Morey
National Institute of Standards and Technology
Standard Reference Data Program
100 Bureau Drive, Stop 2300
Gaithersburg, MD 20899-2300
301-975-2200
website http://www.nist.gov/srd
If there are questions or problems pertaining to the mass spectral data or use of this program, bug fixes, or to suggest improvements, contact:
Dr. Stephen Stein
National Institute of Standards and Technology
100 Bureau Drive, Stop 8362
Gaithersburg, MD 20899-8362
e-mail:
Phone: 1 (301) 975-2505
FAX #: 1 (301) 926-4513
or
O. David Sparkman
5354 Thunderbird Court
Antioch, CA 94531
Phone: 1 (925) 754-5003
Cell: 1 (209) 483-5740
FAX#: 1 (772) 264-6150
For more general questions, ordering, upgrades, and basic support, contact your NIST distributor:
Scientific Instrument Services, Inc.
See http://www.sisweb.com/nist concerning the latest news, upgrades, and support available for NIST MS. NIST sometimes provides free updates and enhancements to the NIST Mass Spectral Search Program and AMDIS. They can be downloaded from our web site. Upgrades to the NIST/EPA/NIH Mass Spectral Library must be purchased.
[1] In upgrading from a previous version, the Database and Programs will be overwritten by default. To avoid this, (1) the Database should be installed in a new folder, and (2) a new Program Folder should be selected for the Start Programs shortcuts.
(1) To retain the previous version of the NIST MS Search Program and its associated Database, select No in the Question dialog box below.
Question dialog box: Select overwriting the previous installation, or retain the previous installation and install NIST 14 in a new folder.
(2) When the dialog box shown below appears, the name of the Program Folder must be changed to NIST 14 Mass Spectral Database. If this is not done, the contents of the existing Program Folder, NIST Mass Spectral Database, will be overwritten, and the previous version of MS Search and its Database will no longer be accessible from the Start menu.
The name of the Program Folder must be changed if a previous version of MS Search and its Database are to be retained.
[2] Library searchable with In-source HiRes search has files peak_em0.inu and peak_em0.dbu
[3] New convenience features added to version 2.0g included:
· Default view of Constraints and Other Searches dialog boxes changed to provide more convenience to the users.
· Ability to Sort the Spec List alphabetically, by number of synonyms, by number of other databases.
· Toggle between Tab views (Lib. Search, Other Search, Name, Compare, Librarian, and MSMS Windows) using Crtl‑Tab or Ctrl-Shift-Tab.
· When a Tag from the “Comment” field is displayed separately, its display in the “Comment” field is suppressed.
[4] The words precursor and parent are used interchangeably in this documentation,
[5] If RI values of multiple specified Column Types are associated with a spectrum, than RI of the first available column type in the following sequence is output into a JCAMP file:
- Semi-Standard Non-Polar
- Standard Non-Polar
- Standard Polar
[6] "MS/MS Spectra only" option disables "Keep IDs unchanged". To override this, use command line option /KeepIDs=YES
[7] In most cases, the "2008 MS Search compatible" option should be turned OFF. This option is intended only for saving MS/MS spectra into a MSP text file in a format, which is compatible with old versions of MS Search. MS/MS or In-source HiRes libraries created with this option have obsolete format unsuitable for reliable searching of high resolution spectra.